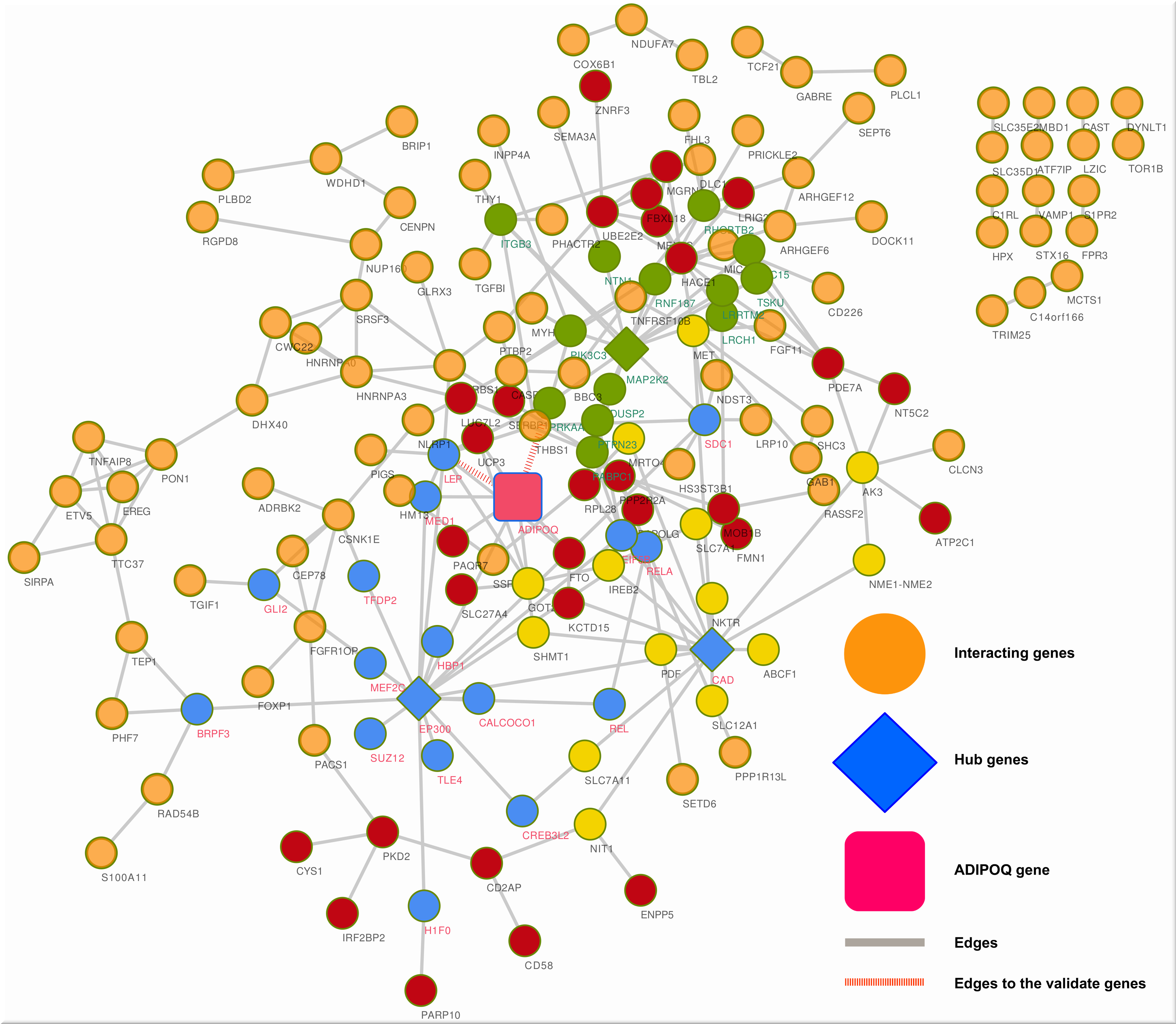
Softwares/tools, Web Server/Databases
We are always giving our findings in the form of software/tools, web server and databases. We also host data repositories of our studies. All developed softwares and tools list below
Software and Webserver
miR-Hy : A robust shallow-deep learning system for plant microRNA discovery
Coming Soon... |
DeepPlnc : A tool to detect plant lncRNAs through multimodal deep learning on sequential data
Coming Soon... |
|
RBPSpot : A tool to identify RBP binding spots in RNA 
Related article for citation: Read research article here Accepted 
Standalone version of the program: Github link |
miRbiom : A webserver for miRNA profiling and miRNA biogenesis-a resultant of spatio-temporal combinations of several RNA-binding proteins. Cite: PLoS ONE, 2021
 Standalone version of the program: Github link 
|
Picro-DB : A comprehensive information database for Picrorhiza kurroa genome and its various genomic components
Cite: Scientific Reports, 2021
 |
PRP: The Plant Regulomics Portal: An ultimate resource for integrated information and study of plant regulatory systems including NGS, Epigenomics, miRNA, Transcription factor and network analysis. Enriched with cutting edge browser and visualization tools, selecting downloading and analytical tools. Cite: Database, 2019
|
IPFIP : Molecular systems based study information portal for Idiopathic Pulmonary Fibrosis (IPFIP) Cite: IPFIP (Scientific Reports, April 2017)
|
MauPIR : Click logo image for redirection of database
Cite: MauPIR (Journal of proteome research; May 10, 2016.) |
AssemblyVAlidator : A tool to detect misassembly in de novo assembled contigs Cite: AssemblyVAlidator (Journal of Biosciences; September, 2016)
|
The Mythology of "micro"-RNAs. Click logo image for redirection of database Cite: The Mythology of "micro"-RNAs (Nucleic Acids Research, 2015, doi: 10.1093/nar/gkv871)
|
pPromotif: A novel and accurate algorithm to identify Transcription Factor Binding Sites on plant genomic sequences. Cite:pPromotif (J Biosci June 2014. PMID 24845501.)
|
miReader: New version of miReader coming soon
Cite: Click here for the PLoS ONE paper. Download standalone version of miReader |
miR-BAG: An Avant-garde parallely coded tool, based on machine learning for accurate miRNA identification. Works universely on genomic and NGS (Next Generation Sequencing) data.
Cite: Click here for the PLoS ONE paper. Click logo to access webserver of miR-BAG Download standalone version of miR-BAG |
p-TAREF:
A concurrent application for fast and accurate plant miRNA target identifications.
Cite: Click here fore the paper. BMC Genomics (2011), 12:636. * (Compatible browsers: Firefox, Google Chrome) Click logo to download standalone verion of P-TAREF 
|
filteR: Parallely coded tool for fast Next Generation Sequencing Read filtering and cleaning. (Coded in Qt, C++, Parallel library used:OpenMP). Download here the standalone verion Cite: Click here for the paper. |
TAREF: A tool to Refine microRNAs target prediction in animal. Citation: Heikham R and Shankar R, 2010, Journal of Biosciences, 35, 105-118. Click here for papers citing this work. |