
A revolutionary algorithm to identify plant pre-miRNAs
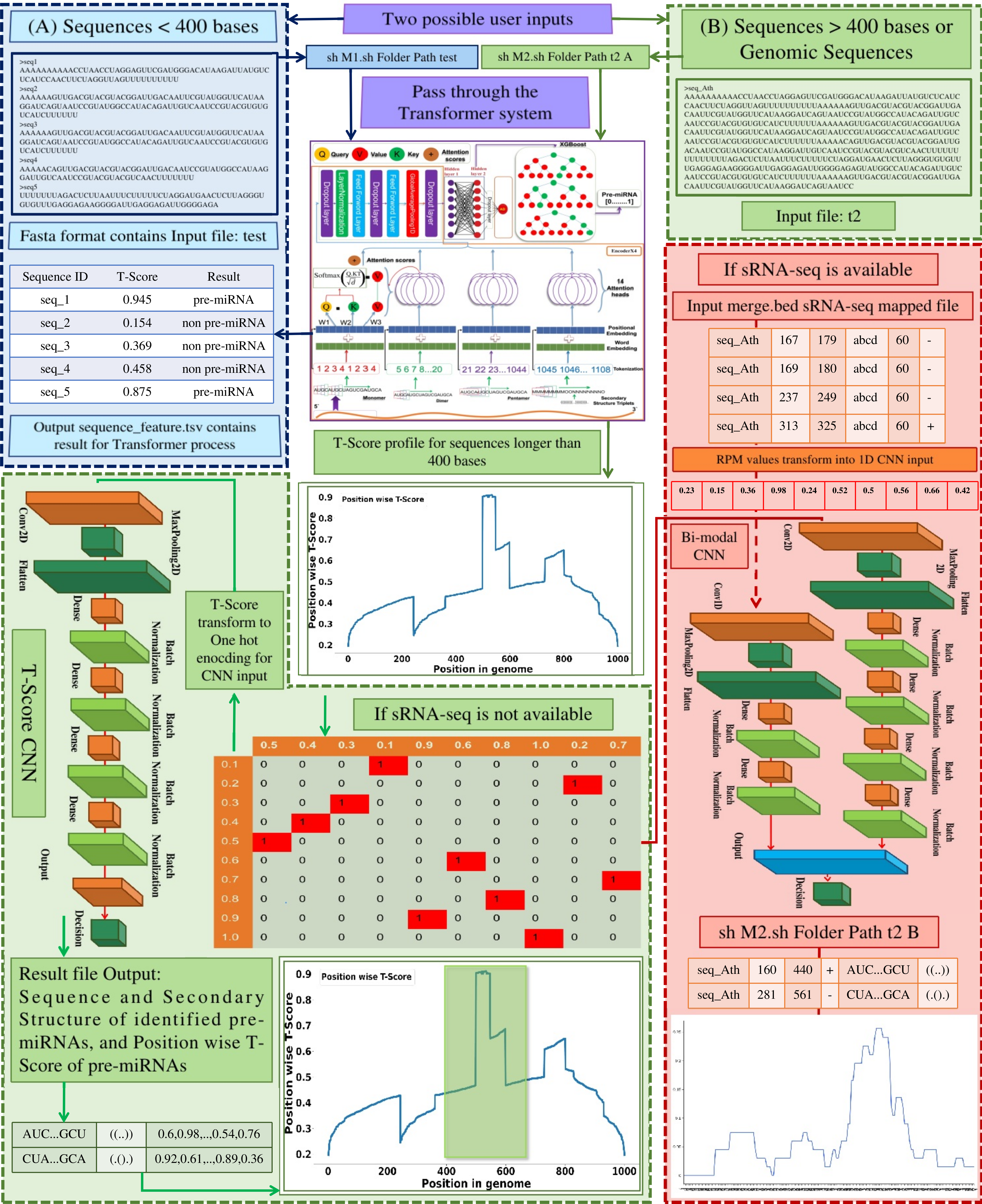
Illustration to running workflow of standalone program
Running the standalone program
To build model implementing hyperparameter tuning
Example: python3 hyper_param.py file_for_tuning
Input file description
file_for_tuning = file containing label (0/1), sequence (sequence containing pre-miRNAs and non-pre-miRNAs), and dot bracket ("(">"M", ".">"O", ")">"N"). All in one line separated by tab for a single instance.
| Label | Sequence | Secondary Structure |
|---|---|---|
| 1 | UGAUAAACAAAGUGUGUAACAUCACCUCAUCUACAUGUGUGAUUUUUUUUUUGAAUAUAGACAACUU UUUAGUCAGAGUUUACAUGAGUUUUCACCUAAUUUGUGGUUUAAUUACACCGCAUAUUUGCCCAAUU UAGUGAGUAUAGUGAGUUUCUGUAGAGAAGCUCAUCUUAGAAUUAUUCAUGUAUUCCACUACUAAA |
OOOOOOOOMMMMMMMOMMOOMMMMMOOMMMOOOONNNONNNNNOOOOOOOOOOOOOONNONNONNNN NMMMMMOMMOMOOMMMMMMMMMOMMMOOOOOOOOOMMMMMOOOOOOOONNNNNMMMMMMMMOOOOOO OONNNNNNNNOMMMMMMMMMMOOOONNNNNNNNNNOOOONNNOONNNNNNNNNOONOONNNNNNNO |
| 0 | AGAUCUACAAGAGAAGAUAAGUUUGAGGCAAAUUCGAGAUCUGGAAGCUGGUUUUCUCUUUACAAAU AACACUAACCCUACCAUCAAAUCAAGAAAGGAGGCUUUGAACAAAUAGCUUGAUUGAAGUAUGAAGU GGCUCGGUGGGCGACGAUGACGGGCGAGCUCCGGCGAGGGCCUGGGGGCCUGAGCGACGAUGACGG | OOMMMOOOMMMMMMMMMMOMMMMMOMMMMOOOOOOOONONNNOONNNNNONNNNNNNNNNOOOOOOO OOMMMMOOOMOMMMOOMMMMOMMMMMMMMMOOOONNNNOOOOOOOOOONNNNNNNNNONNNONONNN NMMMMMOMOMOOOOOOOOOONONONNNNNOOOOMMOOMMMMMOOOONNNNNOONNOOONNNOOOOO |
Output file description
param.txt = Optimized hyparameters for Transformers.
| Name | Value |
|---|---|
| learning_rate | 0.583 |
| activation | relu |
| activation2 | selu |
| activation3 | LeakyRelu |
| batch_size | 40 |
| embed_dim | 28 |
| epochs | 20 |
| num_heads | 14 |
| ff_dim | 14 |
| neurons | 38 |
| neurons2 | 12 |
| dropout_rate | 0.16 |
| dropout_rate2 | 0.17 |
| Optimizer | Adadelta |
miWords.sav = Hyperparameter optimized trained model for XGBoost.
Module 1: Sequence having length <= 400 bases
Example: sh M1.sh folderpath test
Output description
sequence_feature.tsv = Classification result of the sequence provided.
| Sequence ID | T-Score | Result |
|---|---|---|
| seq_1 | 0.945 | pre-miRNA |
| seq_2 | 0.154 | non pre-miRNA |
| seq_3 | 0.389 | non pre-miRNA |
| seq_4 | 0.458 | non pre-miRNA |
| seq_5 | 0.875 | pre-miRNA |
Module 2 (A): Genomic sequence longer than 400 bases (Implementing Transformers + T-Score CNN when sRNA-seq data is not available)
Example: sh M2.sh folderpath genomic_sequence A (genomic_sequences is written as t2 in guide figure.)
Output description
merge.txt = output of module 2 (Sequence, Secondary Struture, Position wise T-Score).
| S. No. | Sequence | Secondary structure | Position wise T-Score |
|---|---|---|---|
| 1 | GCTAGTAAATTTGTTGATTCATGCTTGTAGATGTACACACCACAGCAGATGCATGATGCATGGTTGGCATAGATAAGAATATAGAGGCAGGGGCA AAGGAAGCATGAGCTTGTGGGAGTTGTAGGAATGGATGCGTAGAGAAAGAAATGAAATCTGAACCAGA TTTGTGATTTGATTCTTTCCTATGACGTCCATTCCAATGATTTCCAGTCGCTCCTTCCTTGACCCAAACTGCTTCTTCTCTCCCCATTCCCCAAGGATGCAGCCAAATTACTCCTAA | ...(((((.((.((((.......(((((..((((.((.((((..(((.........))).))))))))))..))))).....((((((( ((((..(((((((...((((...((((((((((.(((((((((((((((.(((((((.(.(((((.............. ))))).))))))))))))).)))))))))).))))))))))...)))))))))))..)))...))))))))........(((((.....))))))))).)).)))))..... | [0.92965674, 0.07397337, 0.07397337, 0.07397337, 0.22203702, 0.92965674] |
genomic_sequence.bed = output from T-Score CNN (bed6 format).
| Sequence ID | Start | End | Name | Score | Strand |
|---|---|---|---|---|---|
| seq_Ath | 387 | 667 | abcd | 60 | + |
| seq_Ath | 407 | 687 | abcd | 60 | - |
seq.txt = Sequence position window wise T-score of the sequence region in the window.
| Sequence ID | Position | T-Score | Sequence | Secondary structure |
|---|---|---|---|---|
| seq_Ath | 0 | 0.07397337 | AAUUAGUCGUAUUCAGUUGUAAAUUCGUUUUUCGUGUAAUGGAGGGAGUACAGGACAAGCUGGCAAGUGGUCUUUGGAUUCCAUGAAGCCUGCUGCCGCGUACAGAAGUCACUAGUGUAGUAGUGGCA CUGAAACGACGUGUGCAUGCUGAUGCUGUCGCCGCAUCCCAUCCCAGUCCUUUUUUUUUUCGGGGACCCAUC | .....(((((.((((((..........((((((........))))))((((.((.((.((.(((..((((((....))..))))...))).))))))..))))....(((((((......)))))))) )))))))))).((.(.(((..(((((.......))))).))).)))((((((.........))))))..... |
| seq_Ath | 1 | 0.07397337 | AUUAGUCGUAUUCAGUUGUAAAUUCGUUUUUCGUGUAAUGGAGGGAGUACAGGACAAGCUGGCAAGUGGUCUUUGGAUUCCAUGAAGCCUGCUGCCGCGUACAGAAGUCACUAGUGUAGUAGUGGCAC UGAAACGACGUGUGCAUGCUGAUGCUGUCGCCGCAUCCCAUCCCAGUCCUUUUUUUUUUCGGGGACCCAUCC | ....(((((.((((((..........((((((........))))))((((.((.((.((.(((..((((((....))..))))...))).))))))..))))....(((((((......))))))))) ))))))))).((.(.(((..(((((.......))))).))).)))((((((.........))))))...... |
| seq_Ath | 2 | 0.07397337 | UUAGUCGUAUUCAGUUGUAAAUUCGUUUUUCGUGUAAUGGAGGGAGUACAGGACAAGCUGGCAAGUGGUCUUUGGAUUCCAUGAAGCCUGCUGCCGCGUACAGAAGUCACUAGUGUAGUAGUGGCACU GAAACGACGUGUGCAUGCUGAUGCUGUCGCCGCAUCCCAUCCCAGUCCUUUUUUUUUUCGGGGACCCAUCCC | ...(((((.((((((..........((((((........))))))((((.((.((.((.(((..((((((....))..))))...))).))))))..))))....(((((((......)))))))))) )))))))).((.(.(((..(((((.......))))).))).)))((((((.........))))))....... |
| seq_Ath | 3 | 0.07397337 | UAGUCGUAUUCAGUUGUAAAUUCGUUUUUCGUGUAAUGGAGGGAGUACAGGACAAGCUGGCAAGUGGUCUUUGGAUUCCAUGAAGCCUGCUGCCGCGUACAGAAGUCACUAGUGUAGUAGUGGCACUG AAACGACGUGUGCAUGCUGAUGCUGUCGCCGCAUCCCAUCCCAGUCCUUUUUUUUUUCGGGGACCCAUCCCA | ..(((((.((((((..........((((((........))))))((((.((.((.((.(((..((((((....))..))))...))).))))))..))))....(((((((......))))))))))) ))))))).((.(.(((..(((((.......))))).))).)))((((((.........))))))........ |
merge = folder containing "csv" files to construct line plot overlapping sequence.
Module 2 (B): If the user is providing sequencing read data (sRNA-seq) and also sequence length is >400 bases:
User needs to process the fastq file into bed6 format by instructions mentioned below (Note: User can select their own mapping software)
Step 1: Build Genome Index: hisat2-build genome index
Step 2: Read Mapping: hisat2 -x index -U read.fastq -S read.sam
Step 3: Convert SAM to bed6 format: sam2bed read.sam |cut -f1-6 > read.bed
If multiple condition are available then merge them into one bed file: cat *.bed >read.bed
Step 4: sh M2.sh folderpath genomic_sequence B read.bed (genomic_sequences is written as t2 in guide figure.)
Output description
read.bed = read alignment data in bed6 format (converted from SAM) (generated by user).
| Sequence ID | Start | End | Name | Score | Strand |
|---|---|---|---|---|---|
| seq_Ath | 167 | 179 | abcd | 60 | - |
| seq_Ath | 169 | 180 | abcd | 60 | - |
| seq_Ath | 237 | 249 | abcd | 60 | - |
| seq_Ath | 313 | 325 | abcd | 60 | + |
| seq_Ath | 535 | 549 | abcd | 60 | + |
| seq_Ath | 542 | 553 | abcd | 60 | - |
| seq_Ath | 542 | 553 | abcd | 60 | - |
| seq_Ath | 542 | 553 | abcd | 60 | - |
| seq_Ath | 542 | 553 | abcd | 60 | - |
| seq_Ath | 542 | 554 | abcd | 60 | - |
genomic_sequence = Fasta sequence utilized for Bimodal CNN.
genomic_sequence.bed = output from Bimodal CNN (bed6 format).
| Sequence ID | Start | End | Name | Score | Strand |
|---|---|---|---|---|---|
| seq_Ath | 91 | 371 | abcd | 60 | + |
| seq_Ath | 406 | 686 | abcd | 60 | + |
| seq_Ath | 435 | 715 | abcd | 60 | - |
| seq_Ath | 521 | 801 | abcd | 60 | - |
genomic_sequence_rpm.txt = Final result (ID, Start, End, Strand, Sequence, Secondary Struture).
| Sequence ID | Start | End | Strand | Sequence | Secondary structure |
|---|---|---|---|---|---|
| seq_Ath | 397 | 677 | + | UUGUUGAUUCAUGCUUGUAGAUGUACACACCACAGCAGAUGCAUGAUGCAUGGUUGGCAUAGAUAAGAAUAUAGAGGCAGGGGCAAAGGAAGCAUGAGCUUG UGGGAGUUGUAGGAAUGGAUGCGUAGAGAAAGAAAUGAAAUCUGAACCAGAUUUGUGAUUUGAUUCUUUCCUAUGACGUCCAUUCCAAUGAUUUCCAGUCGCUCCUUCCUUGACCCAAACUGCUUCUU CUCUCCCCAUUCCCCAAGGAUGCAGCCAAAUUACUCCUAAUUUGCUCCUC | .(((((((.(((((.((((..(((..........)))..))))....))))))))))))......(((....(((((((((((..(((((((...((((...((((((((((.(((((((((((((((.(((((((. (.(((((..............))))).))))))))))))).)))))))))).))))))))))...)))))))))))..)))...))))))))..)))............((((.(((....(((((....)))))))))))). |
| seq_Ath | 407 | 687 | - | GGCCGACCUGGAGGAGCAAAUUAGGAGUAAUUUGGCUGCAUCCUUGGGGAAUGGGGAGAGAAGAAGCAGUUUGGGUCAAGGAAGGAGCGACUGGAAAUCAUU GGAAUGGACGUCAUAGGAAAGAAUCAAAUCACAAAUCUGGUUCAGAUUUCAUUUCUUUCUCUACGCAUCCAUUCCUACAACUCCCACAAGCUCAUGCUUCCUUUGCCCCUGCCUCUAUAUUCUUAUCU AUGCCAACCAUGCAUCAUGCAUCUGCUGUGGUGUGUACAUCUACAAGCAU | (((...(((.(((((((((((((....)))))....))).))))).))).((((((((((.(((.((((...((((.(((((((((((...(((........((((((((.((..((((((((((........ .(((((((...)))))))...))))))).))).)).))))))))........)))...))))...))))))).)))))))).)))...))))).))))))))....(((((...))))).((((((((((....)).)))).)))). |

>seq_1
UGAUAAACAAAGUGUGUAACAUCACCUCAUCUACAUGUGUGAUUUUUUUUUUGAAUAUAGACAACUUUUUAGUCAGAGUUUACAUGAGUUUUCACCUAAUUUGUGGUUUAAUUACACCGCAUAUUU
GCCCAAUUUAGUGAGUAUAGUGAGUUUCUGUAGAGAAGCUCAUCUUAGAAUUAUUCAUGUAUUCCACUACUAAA
>seq_2
AGAUCUACAAGAGAAGAUAAGUUUGAGGCAAAUUCGAGAUCUGGAAGCUGGUUUUCUCUUUACAAAUAACACUAACCCUACCAUCAAAUCAAGAAAGGAGGCUUUGAACAAAUAGCUUGAUUGAAG
UAUGAAGUGGCUCGGUGGGCGACGAUGACGGGCGAGCUCCGGCGAGGGCCUGGGGGCCUGAGCGACGAUGACGG
>seq_3
AACGGGUCGUGCCGGCACGGCCCACGAGCGGGCGUGCCGUGCCGUUCCUGGGCCGGCUACAGUGCUGCCGUGCUCGGGCCGGCACGCCUUGGCCCGGCCCAUUUGGCCAGGUAUACUAGUCGGCUC
CAGUCCUCCUCCCCCCAGCGACCUAAGCCGCCACCGCCCUCGCCGCGCUACCGCCAGCGCCGCCUGCCGUCCCU
>seq_4
UUUUCCCCUUGAUUUUAGGGUUAGGGUUUCAUGAUUUGGGGAAAAAUUUGGGAUCUUACUGUAGCUAGGGUUUCGGUUCUUGGGGAUUUGUCUGAGAUUUGCAUGAACUUUUGCUUUCCCCUUCUU
CUAAUUUCUUCAACCCGAAACCCUAGAAGGCCUAAUUCCAUUUCUUAUAUUUCGGGAUUGCAUGAUUUGGGCUU
>seq_5
CCGGACGAUAGCAAGCGCUGGCAGUAGAGUAGGCUAGAGUCAUGAGUCUGAGUCAUGCUGGCUUUAUAUAGACAAAAAAUGGUACUACACACAAAUGAAAUUUCUAGCAAAAAUAAUCAAUGCACU
UUCCUUGAUUACACACCAACUUUAUGUAUAUAUAGGCUGGAAUAAUCCAUUGUGCAUGUACAUGAAUAUAGAUU
>seq_Ath
UGAUAAACAAAGUGUGUAACAUCACCUCAUCUACAUGUGUGAUUUUUUUUUUGAAUAUAGACAACUUUUUAGUCAGAGUUUACAUGAGUUUUCACCUAAUUUGUGGUUUAAUUACACCGCAUAUUU
GCCCAAUUUAGUGAGUAUAGUGAGUUUCUGUAGAGAAGCUCAUCUUAGAAUUAUUCAUGUAUUCCACUACUAAAAGAUCUACAAGAGAAGAUAAGUUUGAGGCAAAUUCGAGAUCUGGAAGCUGGU
UUUCUCUUUACAAAUAACACUAACCCUACCAUCAAAUCAAGAAAGGAGGCUUUGAACAAAUAGCUUGAUUGAAGUAUGAAGUGGCUCGGUGGGCGACGAUGACGGGCGAGCUCCGGCGAGGGCCUG
GGGGCCUGAGCGACGAUGACGGAACGGGUCGUGCCGGCACGGCCCACGAGCGGGCGUGCCGUGCCGUUCCUGGGCCGGCUACAGUGCUGCCGUGCUCGGGCCGGCACGCCUUGGCCCGGCCCAUUU
GGCCAGGUAUACUAGUCGGCUCCAGUCCUCCUCCCCCCAGCGACCUAAGCCGCCACCGCCCUCGCCGCGCUACCGCCAGCGCCGCCUGCCGUCCCUUUUUCCCCUUGAUUUUAGGGUUAGGGUUUC
AUGAUUUGGGGAAAAAUUUGGGAUCUUACUGUAGCUAGGGUUUCGGUUCUUGGGGAUUUGUCUGAGAUUUGCAUGAACUUUUGCUUUCCCCUUCUUCUAAUUUCUUCAACCCGAAACCCUAGAAGG
CCUAAUUCCAUUUCUUAUAUUUCGGGAUUGCAUGAUUUGGGCUUCCGGACGAUAGCAAGCGCUGGCAGUAGAGUAGGCUAGAGUCAUGAGUCUGAGUCAUGCUGGCUUUAUAUAGACAAAAAAUGG
UACUACACACAAAUGAAAUUUCUAGCAAAAAUAAUCAAUGCACUUUCCUUGAUUACACACCAACUUUAUGUAUAUAUAGGCUGGAAUAAUCCAUUGUGCAUGUACAUGAAUAUAGAUU
B: Bimodal CNN
@SEQ_ID
GATTTGGGGTTCAAAGCAGTATCGATCAAATAGTAAATCCATTTGTTCAACTCACAGTTT
+
!''*((((***+))%%%++)(%%%%).1***-+*''))**55CCF>>>>>>CCCCCCC65
chr1 0 100 abcd 255 +
chr1 10 125 abcd 255 -

