
A revolutionary algorithm to identify plant pre-miRNAs
About miWords:
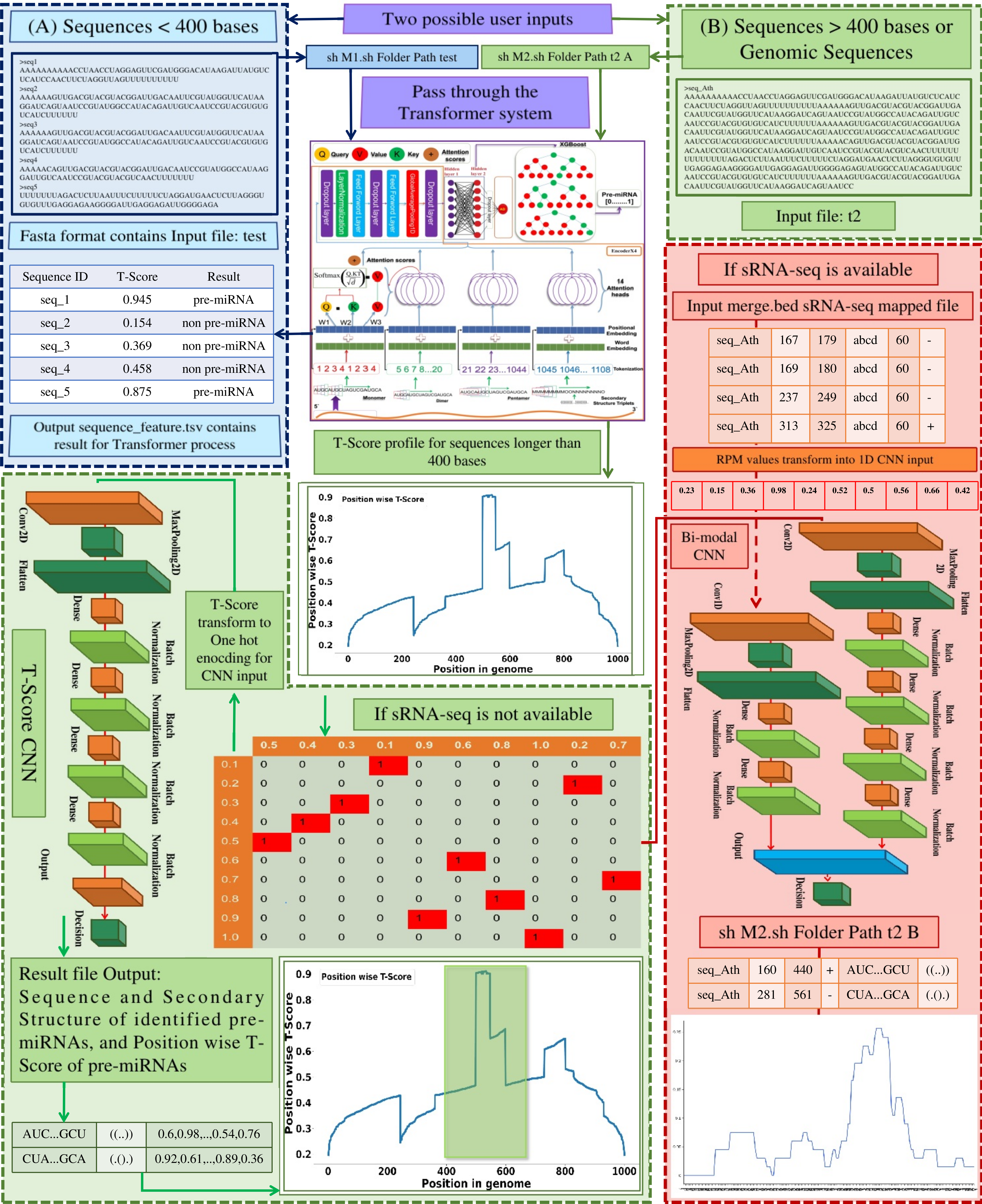
Discovering pre-miRNAs is the core of miRNA discovery. Using ...traditional sequence/structural features many tools have been published to discover miRNAs. However, in practical applications like genomic annotations, their actual performance has been very low. This becomes more grave in plants where unlike animals pre-miRNAs are much more complex and difficult to identify. A huge gap exists between animals and plants for the available software for miRNA discovery and species specific miRNAs information. Here, we present miWords, a composite deep-learning system of transformers and CNNs which sees genome as a pool of sentences made of words with specific occurrence preferences and contexts, to accurately identify pre-miRNA regions across plant genomes. A comprehensive bench-marking was done involving >10 software representing different genre and many experimentally validated datasets. miWords emerged as the best one while breaching accuracy of 98% and performance lead of ~10%. miWords was also evaluated across Arabidopsis genome where also it outperformed the compared tools. As a demonstration, miWords was run across the Tea genome, reporting 803 pre-miRNA regions, all validated by sRNA-seq reads from multiple samples and most of them were functionally supported by the degradome sequencing data. miWords is freely available as stand-alone source codes at https://scbb.ihbt.res.in/miWords/index.php.
Notification:
In order to run the complete miWords system (Transformers + T-Score + RPM-CNN), you are encouraged to download and run the standalone code from here.
Just to see how the transformers only part of miWords performs on some sequences (<= 400 bases), please run the below given online program.
Sagar Gupta, Ravi Shankar* Briefings in Bioinformatics, 2023 Read the research article here