Supplementary Data
Table S1: List of some published tools for Transcription factor binding site identification.
Sheet 1: List of some published tools for Transcription factor binding site identification
| S.No. | Software | Algorithm | Encoding scheme | Biological relevance | Dataset | Species | Year | Webserver (W)/ Standalone (S) |
|---|---|---|---|---|---|---|---|---|
| 1 | KmerHMM | HMM | kmer | Model the dependence between adjacent nucleotide positions | PBM | Human | 2013 | W/S |
| 2 | GkmSVM | SVM | gapped-kmer | Detection and modulation of functional sequence elements in regulatory DNA | ChIP-seq | Human | 2014 | S |
| 3 | pPromotif | Probabilistic modeling | position weight matrix, conservation index (Ci Value), and inter-nucleotide dependence | Plant transcription factor binding sites | AGRIS database and TBFS annotations in GenBank entries | Arabidopsis thaliana | 2014 | S |
| 4 | DeepSEA | CNN | One-hot encoding | Identify the noncoding-variant effects de novo from sequence on chromatin | ChIP-seq | Human | 2015 | S |
| 5 | DeepBind | CNN | One-hot encode | Nucleic acid binding site prediction and can discover new patterns even when the locations of patterns within sequences are unkonwn | ChIP-seq | Human | 2015 | S |
| 6 | Basset | CNN | One-hot encode | Learn the complex code of DNA accessibility across many cell types and and annotate every mutation in the genome with its influence on present accessibility and latent potential for accessibility | DNase-seq | Human | 2016 | S |
| 7 | DanQ | CNN and Bi-LSTM | One-hot encode | Identify non-coding function de novo from sequence which can have enormous benefit for both basic science and translational research | ChIP-seq, DNase-seq | Human | 2016 | S |
| 8 | LS-GKM | SVM | Gapped kmer | Identify and detect the regulatory vocabulary encoded in functional DNA elements, and significantly contribute to understand gene regulation. | ChIP-seq | Human | 2016 | S |
| 9 | DeeperBind | CNN+LSTM | One-hot encoding | Model the positional dynamics of probe sequences and hence reckons with the contributions made by individual sub-regions in DNA sequences | PBMs | Human | 2017 | S |
| 10 | DeepSNR | CNN and DeepCNN | One-hot encoding | Identify TF binding location at Single Nucleotide Resolution de novo from DNA sequence and learns the dependencies between nucleotides at different positions within the binding site description | ChIP-exonuclease | Human | 2017 | S |
| 11 | Tfimpute | CNN | One-hot encoding | Identify cell-specific TF binding | ChIP-seq | Human | 2017 | S |
| 12 | KEGRU | Bi-GRU | k-mer embedding | Capture complex context information from the k-mer sequence | ChIP-seq | Human | 2018 | S |
| 13 | DeFine | CNN | One-hot encoding | Identifies cell type-specific functional impact of abundant non-coding variants including SNPs and indels. Also identifies the causal functional non-coding variants from disease-associated variants in GWAS | ChIP-seq | Human | 2018 | W/S |
| 14 | K-mer grammar | Logistic regression | k-mers | Framework to exploit characteristic chromatin contexts and sequence organization to classify regulatory regions based on sequence features - k-mers | ChIP-seq, Mnase-seq | Zea mays | 2019 | S |
| 15 | FactorNet | CNN+BiLSTM | One-hot encoding | Identify cell type-specific transcription factor binding by leveraging signal data, such as DNase I cleavage | ChIP-seq | Human | 2019 | S |
| 16 | DESSO | CNN | One-hot encoding | Identify motifs and identify TFBSs in both sequence and regional DNA shape features | ChIP-seq | Human | 2019 | S |
| 17 | DeepRAM | CNN/RNN | One-hot/k-mer embedding | Uses Different architectures using CNNs or RNNs to identify DNA/RNA sequence binding specificity | ChIP-seq | Human | 2019 | S |
| 18 | WSCNNLSTM | Multi-instance learning and hybrid neural network | k-mer embedding | Identify in-vivo protein-DNA binding | ChIP-seq | Human | 2019 | S |
| 19 | TbiNet | CNN, Bi-LSTM, and Attention mechanism | One-hot encoding | Identify transcription factor binding sites | ChIP-seq | Human | 2020 | S |
| 20 | FCNA | CNN | One-hot encoding | Accurately identify TF-DNA binding motifs across different cell lines and infer indirect TF-DNA bindings | ChIP-seq | Human | 2021 | S |
| 21 | DLBSS | CNN/RNN | One-hot encoding and shape features | Identify TF-DNA binding preference using input DNA sequences and shape properties | PBM | Human | 2021 | S |
| 22 | BPNet | CNN | One-hot encoding | Discove relevant motifs and syntax rules underlying the cis-regulatory code | ChIP–nexus | Human | 2021 | S |
| 23 | SAResNet | Self-attention mechanism+residual network | One-hot encoding | Identify DNA-protein binding and learning of the long-range dependencies from the DNA sequence | ChIP-seq | Human | 2021 | S |
| 24 | SeqConv | CNN | One-hot encoding | Identify more precise TF-DNA interaction regions in plants | ChIP-seq | Zea mays | 2021 | S |
| 25 | TSPTFBS | CNN | One-hot encoding | TFBS prediction in plants | DAP-seq | Arabidopsis thaliana | 2021 | S |
| 26 | DNABERT | BERT | k-mer encoding | Enables direct visualization of nucleotide-level importance and semantic relationship within input sequences for better interpretability and accurate identification of conserved sequence motifs and functional genetic variant candidates. | ChIP-seq | Human | 2021 | S |
| 27 | D-AEDNet | Deep Attentive Encoder-Decoder Neural Network | One-hot encoding | Identify the location of TFs–DNA binding sites in DNA sequences at base-pair level by leveraging the nucleotide position information | ChIP-exo and ChIP-seq | Human | 2021 | S |
| 28 | DeepGRN | CNN+Bi-LSTM+Attention mechanism | One-hot encoding | Automatically and effectively predict transcription factor binding sites | ChIP-seq, DNase-Seq | Human | 2021 | S |
| 29 | Wimtrap | XGBoost | PWM | Identify condition- or organ-specific cis-regulatory elements and TF gene targets, with a great flexibility regarding the input data | ChIP-seq | Arabidopsis thaliana | 2022 | S |
| 30 | PlantBind | CNN+Bi-LSTM | One-hot encoding | Identify potential TFBSs of multiple TFs simultaneously | ChIP-seq | Arabidopsis thaliana | 2022 | S |
| 31 | MAResNet | Top-down and bottom-up attentation mechanism+residual network | One-hot encoding | Identify transcription factor binding sites in DNA sequences | ChIP-seq | Human | 2022 | S |
| 32 | FCNsignal | Encoder + Decoder + Skip Architecture | One-hot encoding | (i)Identify base-resolution signals of binding regions, (ii) discriminating binding or non-binding regions, (iii) locating TF-DNA binding regions, and (iv) Identify binding motifs. | ChIP-seq and ATAC-seq | Human | 2022 | NA |
| 33 | TSPTFBS2.0 | DenseNet CNN | One-hot encoding | TFBS prediction in plants | DAP-seq | Arabidopsis thaliana | 2023 | S |
Table S2: Sheet 1 has details for various ChIP-Seq experiments and DAP-seq experiments for the studied TFs. The second sheet provides details on ten fold random independent trails details. Sheet 3 provides details on Hyperparameters for the transformers of PTFSpot. Sheets 4 and 5 cover the prime motifs details for Datasets "A" and "B", respectively. Sheets 6-8 cover details on experimentally reported motifs. Sheet 9 covers the details of docking study.
Sheet 1: Information of collected data for ChIP and DAP-Seq
| S.No. | Name | Number of Peaks | Source | Assay Type | TF Family |
|---|---|---|---|---|---|
| 1 | ABF1 | 22241 | PCBase | ChIP-Seq | bZIP |
| 2 | ABF3 | 25687 | PCBase | ChIP-Seq | bZIP |
| 3 | ABF4 | 31851 | PCBase | ChIP-Seq | bZIP |
| 4 | ABI5 | 2798 | PCBase | ChIP-Seq | bZIP |
| 5 | ANAC032 | 22315 | PCBase | ChIP-Seq | NAC |
| 6 | ANAC102 | 9476 | PCBase | ChIP-Seq | NAC |
| 7 | AP1 | 8146 | PCBase | ChIP-Seq | MIKC_MADS |
| 8 | AP2 | 329 | PCBase | ChIP-Seq | AP2 |
| 9 | AZF1 | 4783 | PCBase | ChIP-Seq | ZNF |
| 10 | BBM | 17253 | PCBase | ChIP-Seq | AP2 |
| 11 | BZIP28 | 1102 | PCBase | ChIP-Seq | bZIP |
| 12 | CCA1 | 1206 | PCBase | ChIP-Seq | MYB_related |
| 13 | CRY2 | 1812 | PCBase | ChIP-Seq | CRY2 |
| 14 | DELLA | 1650 | PCBase | ChIP-Seq | GRAS |
| 15 | ERF115 | 11318 | PCBase | ChIP-Seq | AP2EREBP |
| 16 | DREB2A | 3797 | PCBase | ChIP-Seq | ERF |
| 17 | FBH3 | 3384 | PCBase | ChIP-Seq | bHLH |
| 18 | FHY1 | 1117 | PCBase | ChIP-Seq | HAD |
| 19 | FHY3 | 444 | PCBase | ChIP-Seq | FAR1 |
| 20 | FIE | 7787 | PCBase | ChIP-Seq | PRC2 |
| 21 | FLM | 2599 | PCBase | ChIP-Seq | MADS |
| 22 | GBF2 | 31090 | PCBase | ChIP-Seq | bZIP |
| 23 | HAT22 | 31291 | PCBase | ChIP-Seq | HD-ZIP |
| 24 | HB5 | 29929 | PCBase | ChIP-Seq | HD-ZIP |
| 25 | HB7 | 22784 | PCBase | ChIP-Seq | HD-ZIP |
| 26 | HBI | 16 | PCBase | ChIP-Seq | bHLH |
| 27 | HSFA1A | 4374 | PCBase | ChIP-Seq | HSF |
| 28 | HSFA6A | 8898 | PCBase | ChIP-Seq | HSF |
| 29 | IBH1 | 576 | PCBase | ChIP-Seq | bHLH |
| 30 | JAG | 1126 | PCBase | ChIP-Seq | C2H2 |
| 31 | KAN1 | 20007 | PCBase | ChIP-Seq | G2like |
| 32 | LEC1 | 16286 | PCBase | ChIP-Seq | NFY |
| 33 | LFY | 10818 | PCBase | ChIP-Seq | LFY |
| 34 | MYB3 | 20813 | PCBase | ChIP-Seq | MYB |
| 35 | MYB3R3 | 2484 | PCBase | ChIP-Seq | MYB |
| 36 | NFYB2 | 38170 | PCBase | ChIP-Seq | NFY |
| 37 | NFYC2 | 21822 | PCBase | ChIP-Seq | NFY |
| 38 | PIF1 | 5491 | PCBase | ChIP-Seq | bHLH |
| 39 | PIF4 | 33969 | PCBase | ChIP-Seq | bHLH |
| 40 | PIF5 | 3378 | PCBase | ChIP-Seq | bHLH |
| 41 | REV | 6167 | PCBase | ChIP-Seq | HD-ZIP |
| 42 | RGA | 17359 | PCBase | ChIP-Seq | GRAS |
| 43 | SOC1 | 2273 | PCBase | ChIP-Seq | MADS |
| 44 | SVP | 4330 | PCBase | ChIP-Seq | MADS |
| 45 | TOC1 | 191 | PCBase | ChIP-Seq | PRR |
| 46 | WRKY33 | 7355 | PCBase | ChIP-Seq | WRKY |
| 47 | ZAT6 | 20754 | PCBase | ChIP-Seq | C2H2 |
| 48 | AT5G04760 | 30418 | PCBase | ChIP-Seq | MYB |
| 49 | GBF3 | 39592 | PCBase | ChIP-Seq | bZIP |
| 50 | HB6 | 40197 | PCBase | ChIP-Seq | HD-ZIP |
| 51 | MYB44 | 34581 | PCBase | ChIP-Seq | MYB |
| 52 | PhyA | 2233 | PCBase | ChIP-Seq | PhyA |
| 53 | RD26 | 36219 | PCBase | ChIP-Seq | NAC |
| 54 | SEP3 | 24925 | PCBase | ChIP-Seq | MIKC_MADS |
| 55 | ABF2 | 2299 | Plant Cistrome Database | DAP-Seq | bZIP |
| 56 | ABI5 | 9233 | Plant Cistrome Database | DAP-Seq | bZIP |
| 57 | ABR1 | 11691 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 58 | Adof1 | 11744 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 59 | AGL15 | 2257 | Plant Cistrome Database | DAP-Seq | MADS |
| 60 | AGL63 | 15745 | Plant Cistrome Database | DAP-Seq | MADS |
| 61 | AIL7 | 3535 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 62 | ANAC004 | 14895 | Plant Cistrome Database | DAP-Seq | NAC |
| 63 | ANAC005 | 7671 | Plant Cistrome Database | DAP-Seq | NAC |
| 64 | ANAC013 | 5744 | Plant Cistrome Database | DAP-Seq | NAC |
| 65 | ANAC016 | 15693 | Plant Cistrome Database | DAP-Seq | NAC |
| 66 | ANAC017 | 15802 | Plant Cistrome Database | DAP-Seq | NAC |
| 67 | ANAC020 | 7906 | Plant Cistrome Database | DAP-Seq | NAC |
| 68 | ANAC028 | 8161 | Plant Cistrome Database | DAP-Seq | NAC |
| 69 | ANAC034 | 6013 | Plant Cistrome Database | DAP-Seq | NAC |
| 70 | ANAC038 | 6176 | Plant Cistrome Database | DAP-Seq | NAC |
| 71 | ANAC042 | 6110 | Plant Cistrome Database | DAP-Seq | NAC |
| 72 | ANAC045 | 17921 | Plant Cistrome Database | DAP-Seq | NAC |
| 73 | ANAC046 | 18233 | Plant Cistrome Database | DAP-Seq | NAC |
| 74 | ANAC047 | 7691 | Plant Cistrome Database | DAP-Seq | NAC |
| 75 | ANAC050 | 21201 | Plant Cistrome Database | DAP-Seq | NAC |
| 76 | ANAC053 | 13096 | Plant Cistrome Database | DAP-Seq | NAC |
| 77 | ANAC055 | 7513 | Plant Cistrome Database | DAP-Seq | NAC |
| 78 | ANAC057 | 10212 | Plant Cistrome Database | DAP-Seq | NAC |
| 79 | ANAC058 | 19850 | Plant Cistrome Database | DAP-Seq | NAC |
| 80 | ANAC062 | 10281 | Plant Cistrome Database | DAP-Seq | NAC |
| 81 | ANAC070 | 24684 | Plant Cistrome Database | DAP-Seq | NAC |
| 82 | ANAC071 | 23644 | Plant Cistrome Database | DAP-Seq | NAC |
| 83 | ANAC075 | 4382 | Plant Cistrome Database | DAP-Seq | NAC |
| 84 | ANAC079 | 2817 | Plant Cistrome Database | DAP-Seq | NAC |
| 85 | ANAC083 | 21927 | Plant Cistrome Database | DAP-Seq | NAC |
| 86 | ANAC087 | 6399 | Plant Cistrome Database | DAP-Seq | NAC |
| 87 | ANAC092 | 10364 | Plant Cistrome Database | DAP-Seq | NAC |
| 88 | ANAC094 | 738 | Plant Cistrome Database | DAP-Seq | NAC |
| 89 | ANAC096 | 13107 | Plant Cistrome Database | DAP-Seq | NAC |
| 90 | ANAC103 | 8702 | Plant Cistrome Database | DAP-Seq | NAC |
| 91 | ANL2 | 8312 | Plant Cistrome Database | DAP-Seq | HB |
| 92 | AREB3 | 4617 | Plant Cistrome Database | DAP-Seq | bZIP |
| 93 | ARF2 | 11654 | Plant Cistrome Database | DAP-Seq | ARF |
| 94 | ASL18 | 6328 | Plant Cistrome Database | DAP-Seq | LOBAS2 |
| 95 | AT1G01250 | 725 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 96 | AT1G12630 | 18211 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 97 | At1g19000 | 29932 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 98 | AT1G19040 | 994 | Plant Cistrome Database | DAP-Seq | NAC |
| 99 | At1g19210 | 22451 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 100 | AT1G20910 | 10683 | Plant Cistrome Database | DAP-Seq | ARID |
| 101 | At1g22810 | 8437 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 102 | AT1G24250 | 2233 | Plant Cistrome Database | DAP-Seq | Orphan |
| 103 | At1g25550 | 15049 | Plant Cistrome Database | DAP-Seq | G2like |
| 104 | AT1G28160 | 10300 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 105 | At1g36060 | 4035 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 106 | AT1G44830 | 1635 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 107 | AT1G47655 | 7940 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 108 | At1g49010 | 28552 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 109 | AT1G49560 | 20976 | Plant Cistrome Database | DAP-Seq | G2like |
| 110 | At1g64620 | 34001 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 111 | At1g68670 | 11676 | Plant Cistrome Database | DAP-Seq | G2like |
| 112 | AT1G69570 | 13762 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 113 | At1g69690 | 995 | Plant Cistrome Database | DAP-Seq | TCP |
| 114 | AT1G71450 | 14277 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 115 | At1g72010 | 4626 | Plant Cistrome Database | DAP-Seq | TCP |
| 116 | AT1G72740 | 21483 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 117 | At1g74840 | 10169 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 118 | AT1G76880 | 491 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 119 | AT1G77200 | 27117 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 120 | At1g78700 | 5147 | Plant Cistrome Database | DAP-Seq | BZR |
| 121 | At2g01060 | 21429 | Plant Cistrome Database | DAP-Seq | G2like |
| 122 | At2g03500 | 12731 | Plant Cistrome Database | DAP-Seq | G2like |
| 123 | AT2G15740 | 3257 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 124 | AT2G20110 | 30396 | Plant Cistrome Database | DAP-Seq | CPP |
| 125 | AT2G20400 | 6113 | Plant Cistrome Database | DAP-Seq | G2like |
| 126 | AT2G28810 | 49623 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 127 | AT2G33550 | 8647 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 128 | At2g33710 | 18654 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 129 | AT2G40260 | 21916 | Plant Cistrome Database | DAP-Seq | G2like |
| 130 | At2g44940 | 5163 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 131 | At2g45680 | 450 | Plant Cistrome Database | DAP-Seq | TCP |
| 132 | At3g04030 | 22876 | Plant Cistrome Database | DAP-Seq | G2like |
| 133 | At3g09600 | 29529 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 134 | AT3G09735 | 7818 | Plant Cistrome Database | DAP-Seq | S1Falike |
| 135 | AT3G10030 | 3648 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 136 | AT3G10113 | 27249 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 137 | AT3G10580 | 5180 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 138 | At3g11280 | 2031 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 139 | AT3G12130 | 12290 | Plant Cistrome Database | DAP-Seq | C3H |
| 140 | At3g12730 | 10750 | Plant Cistrome Database | DAP-Seq | G2like |
| 141 | At3g14180 | 6618 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 142 | AT3G16280 | 13312 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 143 | At3g24120 | 52038 | Plant Cistrome Database | DAP-Seq | G2like |
| 144 | AT3G25990 | 2682 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 145 | AT3G42860 | 2415 | Plant Cistrome Database | DAP-Seq | zfGRF |
| 146 | At3g45610 | 34597 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 147 | AT3G52440 | 30433 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 148 | AT3G60490 | 9000 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 149 | At3g60580 | 11065 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 150 | AT4G00250 | 11654 | Plant Cistrome Database | DAP-Seq | GeBP |
| 151 | At4g01280 | 24503 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 152 | At4g16750 | 15677 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 153 | AT4G18450 | 561 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 154 | At4g18890 | 865 | Plant Cistrome Database | DAP-Seq | BZR |
| 155 | AT4G26030 | 11315 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 156 | At4g31060 | 7514 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 157 | At4g32800 | 3779 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 158 | At4g36780 | 6039 | Plant Cistrome Database | DAP-Seq | BZR |
| 159 | AT4G37180 | 19680 | Plant Cistrome Database | DAP-Seq | G2like |
| 160 | At4g38000 | 11840 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 161 | AT5G02460 | 45371 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 162 | At5g04390 | 9238 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 163 | AT5G05550 | 18951 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 164 | At5g05790 | 3605 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 165 | At5g08330 | 2915 | Plant Cistrome Database | DAP-Seq | TCP |
| 166 | At5g08520 | 12645 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 167 | At5g08750 | 968 | Plant Cistrome Database | DAP-Seq | C3H |
| 168 | At5g18450 | 2405 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 169 | AT5G22990 | 4898 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 170 | AT5G23930 | 5551 | Plant Cistrome Database | DAP-Seq | mTERF |
| 171 | At5g29000 | 4704 | Plant Cistrome Database | DAP-Seq | G2like |
| 172 | AT5G45580 | 42093 | Plant Cistrome Database | DAP-Seq | G2like |
| 173 | At5g47390 | 17465 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 174 | AT5G47660 | 12711 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 175 | At5g52660 | 19533 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 176 | AT5G56840 | 31403 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 177 | At5g58900 | 18957 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 178 | AT5G60130 | 19106 | Plant Cistrome Database | DAP-Seq | ABI3VP1 |
| 179 | AT5G61620 | 19901 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 180 | At5g62940 | 29881 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 181 | AT5G63260 | 18676 | Plant Cistrome Database | DAP-Seq | C3H |
| 182 | At5g65130 | 1992 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 183 | At5g66730 | 7624 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 184 | AT5G66940 | 40237 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 185 | ATAF1 | 9197 | Plant Cistrome Database | DAP-Seq | NAC |
| 186 | AtGRF6 | 6529 | Plant Cistrome Database | DAP-Seq | GRF |
| 187 | ATHB13 | 23232 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 188 | ATHB15 | 6538 | Plant Cistrome Database | DAP-Seq | HB |
| 189 | ATHB18 | 5366 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 190 | ATHB20 | 5911 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 191 | ATHB21 | 22139 | Plant Cistrome Database | DAP-Seq | HB |
| 192 | ATHB23 | 49895 | Plant Cistrome Database | DAP-Seq | ZFHD |
| 193 | ATHB24 | 21428 | Plant Cistrome Database | DAP-Seq | ZFHD |
| 194 | ATHB25 | 49795 | Plant Cistrome Database | DAP-Seq | ZFHD |
| 195 | AtHB32 | 21104 | Plant Cistrome Database | DAP-Seq | ZFHD |
| 196 | ATHB33 | 34347 | Plant Cistrome Database | DAP-Seq | ZFHD |
| 197 | ATHB34 | 30751 | Plant Cistrome Database | DAP-Seq | ZFHD |
| 198 | ATHB40 | 51300 | Plant Cistrome Database | DAP-Seq | HB |
| 199 | ATHB53 | 17471 | Plant Cistrome Database | DAP-Seq | HB |
| 200 | ATHB5 | 46401 | Plant Cistrome Database | DAP-Seq | HB |
| 201 | ATHB6 | 9807 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 202 | ATHB7 | 10616 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 203 | AtIDD11 | 3281 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 204 | ATY13 | 19568 | Plant Cistrome Database | DAP-Seq | MYB |
| 205 | BAM8 | 4589 | Plant Cistrome Database | DAP-Seq | BES1 |
| 206 | BBX31 | 16775 | Plant Cistrome Database | DAP-Seq | Orphan |
| 207 | bHLH10 | 9144 | Plant Cistrome Database | DAP-Seq | bHLH |
| 208 | bHLH122 | 10050 | Plant Cistrome Database | DAP-Seq | bHLH |
| 209 | bHLH28 | 8905 | Plant Cistrome Database | DAP-Seq | bHLH |
| 210 | bHLH31 | 1829 | Plant Cistrome Database | DAP-Seq | bHLH |
| 211 | bHLH80 | 5667 | Plant Cistrome Database | DAP-Seq | bHLH |
| 212 | BIM1 | 647 | Plant Cistrome Database | DAP-Seq | bHLH |
| 213 | BIM2 | 40836 | Plant Cistrome Database | DAP-Seq | bHLH |
| 214 | BOS1 | 2175 | Plant Cistrome Database | DAP-Seq | MYB |
| 215 | BPC1 | 17699 | Plant Cistrome Database | DAP-Seq | BBRBPC |
| 216 | bZIP16 | 8404 | Plant Cistrome Database | DAP-Seq | bZIP |
| 217 | bZIP18 | 14191 | Plant Cistrome Database | DAP-Seq | bZIP |
| 218 | bZIP28 | 1726 | Plant Cistrome Database | DAP-Seq | bZIP |
| 219 | bZIP3 | 8295 | Plant Cistrome Database | DAP-Seq | bZIP |
| 220 | bZIP44 | 2679 | Plant Cistrome Database | DAP-Seq | bZIP |
| 221 | bZIP48 | 5105 | Plant Cistrome Database | DAP-Seq | bZIP |
| 222 | bZIP50 | 22131 | Plant Cistrome Database | DAP-Seq | bZIP |
| 223 | bZIP52 | 16537 | Plant Cistrome Database | DAP-Seq | bZIP |
| 224 | bZIP53 | 17446 | Plant Cistrome Database | DAP-Seq | bZIP |
| 225 | bZIP68 | 9044 | Plant Cistrome Database | DAP-Seq | bZIP |
| 226 | BZR1 | 1018 | Plant Cistrome Database | DAP-Seq | BZR |
| 227 | CAMTA1 | 2776 | Plant Cistrome Database | DAP-Seq | CAMTA |
| 228 | CAMTA5 | 773 | Plant Cistrome Database | DAP-Seq | CAMTA |
| 229 | CBF1 | 18542 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 230 | CBF2 | 14034 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 231 | CBF3 | 12163 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 232 | CBF4 | 29710 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 233 | CDF3 | 22820 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 234 | CDM1 | 4777 | Plant Cistrome Database | DAP-Seq | C3H |
| 235 | CEJ1 | 30321 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 236 | COG1 | 11354 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 237 | CRC | 9178 | Plant Cistrome Database | DAP-Seq | C2C2YABBY |
| 238 | CRF10 | 6410 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 239 | CRF4 | 2482 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 240 | CUC1 | 3109 | Plant Cistrome Database | DAP-Seq | NAC |
| 241 | CUC2 | 5942 | Plant Cistrome Database | DAP-Seq | NAC |
| 242 | CUC3 | 6336 | Plant Cistrome Database | DAP-Seq | NAC |
| 243 | DAG2 | 24338 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 244 | DDF1 | 2395 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 245 | DDF2 | 326 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 246 | DEAR2 | 23238 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 247 | DEAR3 | 12720 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 248 | DEAR5 | 2274 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 249 | DEL2 | 1401 | Plant Cistrome Database | DAP-Seq | E2FDP |
| 250 | dof24 | 27058 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 251 | dof42 | 801 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 252 | dof43 | 19070 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 253 | dof45 | 24304 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 254 | DREB19 | 15090 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 255 | E2FA | 7135 | Plant Cistrome Database | DAP-Seq | E2FDP |
| 256 | EIN3 | 1209 | Plant Cistrome Database | DAP-Seq | EIL |
| 257 | EPR1 | 52997 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 258 | ERF105 | 14787 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 259 | ERF10 | 4811 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 260 | ERF115 | 12101 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 261 | ERF11 | 7672 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 262 | ERF13 | 9556 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 263 | ERF15 | 25902 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 264 | ERF2 | 726 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 265 | ERF38 | 5518 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 266 | ERF3 | 3321 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 267 | ERF4 | 15193 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 268 | ERF5 | 846 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 269 | ERF7 | 17694 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 270 | ERF8 | 12829 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 271 | ESE1 | 1512 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 272 | ESE3 | 9328 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 273 | FAR1 | 957 | Plant Cistrome Database | DAP-Seq | FAR1 |
| 274 | FRS9 | 7750 | Plant Cistrome Database | DAP-Seq | ND |
| 275 | FUS3 | 3266 | Plant Cistrome Database | DAP-Seq | ABI3VP1 |
| 276 | GATA11 | 3149 | Plant Cistrome Database | DAP-Seq | C2C2gata |
| 277 | GATA19 | 202 | Plant Cistrome Database | DAP-Seq | C2C2gata |
| 278 | GATA1 | 970 | Plant Cistrome Database | DAP-Seq | C2C2gata |
| 279 | GATA20 | 17519 | Plant Cistrome Database | DAP-Seq | C2C2gata |
| 280 | GATA4 | 1611 | Plant Cistrome Database | DAP-Seq | C2C2gata |
| 281 | GBF3 | 16034 | Plant Cistrome Database | DAP-Seq | bZIP |
| 282 | GBF5 | 6575 | Plant Cistrome Database | DAP-Seq | bZIP |
| 283 | GBF6 | 9894 | Plant Cistrome Database | DAP-Seq | bZIP |
| 284 | GRF9 | 2374 | Plant Cistrome Database | DAP-Seq | GRF |
| 285 | GT1 | 519 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 286 | GT2 | 17094 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 287 | GT3a | 1318 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 288 | GTL1 | 31474 | Plant Cistrome Database | DAP-Seq | Trihelix |
| 289 | HAT2 | 14941 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 290 | HDG1 | 11647 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 291 | HSF21 | 756 | Plant Cistrome Database | DAP-Seq | HSF |
| 292 | HSF3 | 21814 | Plant Cistrome Database | DAP-Seq | HSF |
| 293 | HSF6 | 1227 | Plant Cistrome Database | DAP-Seq | HSF |
| 294 | HSF7 | 4474 | Plant Cistrome Database | DAP-Seq | HSF |
| 295 | HSFA6B | 34926 | Plant Cistrome Database | DAP-Seq | HSF |
| 296 | HSFB3 | 1171 | Plant Cistrome Database | DAP-Seq | HSF |
| 297 | HSFC1 | 8354 | Plant Cistrome Database | DAP-Seq | HSF |
| 298 | HY5 | 10140 | Plant Cistrome Database | DAP-Seq | bZIP |
| 299 | IDD4 | 12546 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 300 | IDD5 | 9121 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 301 | IDD7 | 4720 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 302 | JKD | 2160 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 303 | LBD13 | 3715 | Plant Cistrome Database | DAP-Seq | LOBAS2 |
| 304 | LBD18 | 20264 | Plant Cistrome Database | DAP-Seq | LOBAS2 |
| 305 | LBD19 | 23977 | Plant Cistrome Database | DAP-Seq | LOBAS2 |
| 306 | LBD23 | 1451 | Plant Cistrome Database | DAP-Seq | LOBAS2 |
| 307 | LBD2 | 24314 | Plant Cistrome Database | DAP-Seq | LOBAS2 |
| 308 | LCL1 | 8370 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 309 | LEP | 546 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 310 | LHY1 | 33488 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 311 | LMI1 | 41115 | Plant Cistrome Database | DAP-Seq | HB |
| 312 | LOB | 1461 | Plant Cistrome Database | DAP-Seq | LOBAS2 |
| 313 | MGP | 3771 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 314 | MP | 26659 | Plant Cistrome Database | DAP-Seq | ARF_ecoli |
| 315 | MS188 | 17081 | Plant Cistrome Database | DAP-Seq | MYB |
| 316 | MYB101 | 10887 | Plant Cistrome Database | DAP-Seq | MYB |
| 317 | MYB107 | 13462 | Plant Cistrome Database | DAP-Seq | MYB |
| 318 | MYB116 | 8961 | Plant Cistrome Database | DAP-Seq | MYB |
| 319 | MYB118 | 4304 | Plant Cistrome Database | DAP-Seq | MYB |
| 320 | MYB119 | 15557 | Plant Cistrome Database | DAP-Seq | MYB |
| 321 | MYB121 | 2067 | Plant Cistrome Database | DAP-Seq | MYB |
| 322 | MYB13 | 2733 | Plant Cistrome Database | DAP-Seq | MYB |
| 323 | MYB27 | 8363 | Plant Cistrome Database | DAP-Seq | MYB |
| 324 | MYB33 | 22513 | Plant Cistrome Database | DAP-Seq | MYB |
| 325 | MYB3R1 | 22319 | Plant Cistrome Database | DAP-Seq | MYB |
| 326 | MYB3R4 | 1965 | Plant Cistrome Database | DAP-Seq | MYB |
| 327 | MYB3R5 | 2706 | Plant Cistrome Database | DAP-Seq | MYB |
| 328 | MYB44 | 15418 | Plant Cistrome Database | DAP-Seq | MYB |
| 329 | MYB49 | 9961 | Plant Cistrome Database | DAP-Seq | MYB |
| 330 | MYB55 | 10051 | Plant Cistrome Database | DAP-Seq | MYB |
| 331 | MYB56 | 6039 | Plant Cistrome Database | DAP-Seq | MYB |
| 332 | MYB57 | 12402 | Plant Cistrome Database | DAP-Seq | MYB |
| 333 | MYB58 | 21702 | Plant Cistrome Database | DAP-Seq | MYB |
| 334 | MYB61 | 17756 | Plant Cistrome Database | DAP-Seq | MYB |
| 335 | MYB62 | 12953 | Plant Cistrome Database | DAP-Seq | MYB |
| 336 | MYB63 | 2666 | Plant Cistrome Database | DAP-Seq | MYB |
| 337 | MYB65 | 12876 | Plant Cistrome Database | DAP-Seq | MYB |
| 338 | MYB67 | 19668 | Plant Cistrome Database | DAP-Seq | MYB |
| 339 | MYB70 | 11564 | Plant Cistrome Database | DAP-Seq | MYB |
| 340 | MYB73 | 9380 | Plant Cistrome Database | DAP-Seq | MYB |
| 341 | MYB74 | 3061 | Plant Cistrome Database | DAP-Seq | MYB |
| 342 | MYB77 | 27263 | Plant Cistrome Database | DAP-Seq | MYB |
| 343 | MYB81 | 3231 | Plant Cistrome Database | DAP-Seq | MYB |
| 344 | MYB83 | 14502 | Plant Cistrome Database | DAP-Seq | MYB |
| 345 | MYB88 | 15118 | Plant Cistrome Database | DAP-Seq | MYB |
| 346 | MYB93 | 7813 | Plant Cistrome Database | DAP-Seq | MYB |
| 347 | MYB96 | 1147 | Plant Cistrome Database | DAP-Seq | MYB |
| 348 | MYB98 | 5558 | Plant Cistrome Database | DAP-Seq | MYB |
| 349 | MYB99 | 4060 | Plant Cistrome Database | DAP-Seq | MYB |
| 350 | NAC2 | 23673 | Plant Cistrome Database | DAP-Seq | NAC |
| 351 | NAM | 7706 | Plant Cistrome Database | DAP-Seq | NAC |
| 352 | NAP | 6299 | Plant Cistrome Database | DAP-Seq | NAC |
| 353 | NGA4 | 2235 | Plant Cistrome Database | DAP-Seq | ABI3VP1 |
| 354 | NLP7 | 867 | Plant Cistrome Database | DAP-Seq | RWPRK |
| 355 | NST1 | 24770 | Plant Cistrome Database | DAP-Seq | NAC |
| 356 | NTL8 | 5913 | Plant Cistrome Database | DAP-Seq | NAC |
| 357 | NTM1 | 15614 | Plant Cistrome Database | DAP-Seq | NAC |
| 358 | NTM2 | 5202 | Plant Cistrome Database | DAP-Seq | NAC |
| 359 | OBP1 | 23458 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 360 | OBP3 | 21664 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 361 | OBP4 | 27993 | Plant Cistrome Database | DAP-Seq | C2C2dof |
| 362 | PDF2 | 2160 | Plant Cistrome Database | DAP-Seq | Homeobox |
| 363 | PHV | 10285 | Plant Cistrome Database | DAP-Seq | HB |
| 364 | PLT3 | 541 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 365 | PUCHI | 2691 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 366 | Rap210 | 12600 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 367 | RAP211 | 16230 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 368 | RAP212 | 6571 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 369 | RAP21 | 2103 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 370 | RAP26 | 8572 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 371 | RAV1 | 14740 | Plant Cistrome Database | DAP-Seq | RAV |
| 372 | REM19 | 40336 | Plant Cistrome Database | DAP-Seq | REM |
| 373 | RKD2 | 10284 | Plant Cistrome Database | DAP-Seq | RWPRK |
| 374 | RRTF1 | 8037 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 375 | RVE1 | 39879 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 376 | SGR5 | 18969 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 377 | SMB | 40054 | Plant Cistrome Database | DAP-Seq | NAC |
| 378 | SND2 | 23978 | Plant Cistrome Database | DAP-Seq | NAC |
| 379 | SND3 | 45691 | Plant Cistrome Database | DAP-Seq | NAC |
| 380 | SOL1 | 21836 | Plant Cistrome Database | DAP-Seq | CPP |
| 381 | SPL13 | 1179 | Plant Cistrome Database | DAP-Seq | SBP |
| 382 | SPL14 | 2529 | Plant Cistrome Database | DAP-Seq | SBP |
| 383 | SPL15 | 19981 | Plant Cistrome Database | DAP-Seq | SBP |
| 384 | SPL1 | 12940 | Plant Cistrome Database | DAP-Seq | SBP |
| 385 | SPL5 | 27217 | Plant Cistrome Database | DAP-Seq | SBP |
| 386 | SPL9 | 41646 | Plant Cistrome Database | DAP-Seq | SBP |
| 387 | SRS7 | 11330 | Plant Cistrome Database | DAP-Seq | SRS |
| 388 | STOP1 | 1356 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 389 | STZ | 36348 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 390 | SVP | 20687 | Plant Cistrome Database | DAP-Seq | MADS |
| 391 | TBP3 | 6449 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 392 | TCP20 | 1167 | Plant Cistrome Database | DAP-Seq | TCP |
| 393 | TCP24 | 3352 | Plant Cistrome Database | DAP-Seq | TCP |
| 394 | TCP3 | 1416 | Plant Cistrome Database | DAP-Seq | TCP |
| 395 | TCP7 | 1002 | Plant Cistrome Database | DAP-Seq | TCP |
| 396 | TCX2 | 24443 | Plant Cistrome Database | DAP-Seq | CPP |
| 397 | TGA10 | 5500 | Plant Cistrome Database | DAP-Seq | bZIP |
| 398 | TGA1 | 12377 | Plant Cistrome Database | DAP-Seq | bZIP |
| 399 | TGA2 | 16587 | Plant Cistrome Database | DAP-Seq | bZIP |
| 400 | TGA3 | 3451 | Plant Cistrome Database | DAP-Seq | bZIP |
| 401 | TGA4 | 10140 | Plant Cistrome Database | DAP-Seq | bZIP |
| 402 | TGA5 | 2604 | Plant Cistrome Database | DAP-Seq | bZIP |
| 403 | TGA6 | 28254 | Plant Cistrome Database | DAP-Seq | bZIP |
| 404 | TGA9 | 18677 | Plant Cistrome Database | DAP-Seq | bZIP |
| 405 | TINY | 1775 | Plant Cistrome Database | DAP-Seq | AP2EREBP |
| 406 | TRP1 | 1042 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 407 | TRP2 | 3036 | Plant Cistrome Database | DAP-Seq | MYBrelated |
| 408 | VIP1 | 1523 | Plant Cistrome Database | DAP-Seq | bZIP |
| 409 | VND1 | 7156 | Plant Cistrome Database | DAP-Seq | NAC |
| 410 | VND2 | 16577 | Plant Cistrome Database | DAP-Seq | NAC |
| 411 | VND3 | 6534 | Plant Cistrome Database | DAP-Seq | NAC |
| 412 | VND4 | 18184 | Plant Cistrome Database | DAP-Seq | NAC |
| 413 | VND6 | 15899 | Plant Cistrome Database | DAP-Seq | NAC |
| 414 | VRN1 | 7028 | Plant Cistrome Database | DAP-Seq | ABI3VP1 |
| 415 | WIP5 | 8396 | Plant Cistrome Database | DAP-Seq | C2H2 |
| 416 | WRKY14 | 13405 | Plant Cistrome Database | DAP-Seq | WRKY |
| 417 | WRKY15 | 29394 | Plant Cistrome Database | DAP-Seq | WRKY |
| 418 | WRKY18 | 30301 | Plant Cistrome Database | DAP-Seq | WRKY |
| 419 | WRKY20 | 2213 | Plant Cistrome Database | DAP-Seq | WRKY |
| 420 | WRKY21 | 1537 | Plant Cistrome Database | DAP-Seq | WRKY |
| 421 | WRKY22 | 37563 | Plant Cistrome Database | DAP-Seq | WRKY |
| 422 | WRKY24 | 18866 | Plant Cistrome Database | DAP-Seq | WRKY |
| 423 | WRKY25 | 20979 | Plant Cistrome Database | DAP-Seq | WRKY |
| 424 | WRKY27 | 23333 | Plant Cistrome Database | DAP-Seq | WRKY |
| 425 | WRKY28 | 13501 | Plant Cistrome Database | DAP-Seq | WRKY |
| 426 | WRKY29 | 18818 | Plant Cistrome Database | DAP-Seq | WRKY |
| 427 | WRKY30 | 4374 | Plant Cistrome Database | DAP-Seq | WRKY |
| 428 | WRKY31 | 3489 | Plant Cistrome Database | DAP-Seq | WRKY |
| 429 | WRKY33 | 5593 | Plant Cistrome Database | DAP-Seq | WRKY |
| 430 | WRKY3 | 1287 | Plant Cistrome Database | DAP-Seq | WRKY |
| 431 | WRKY40 | 7114 | Plant Cistrome Database | DAP-Seq | WRKY |
| 432 | WRKY45 | 13329 | Plant Cistrome Database | DAP-Seq | WRKY |
| 433 | WRKY50 | 13630 | Plant Cistrome Database | DAP-Seq | WRKY |
| 434 | WRKY55 | 9034 | Plant Cistrome Database | DAP-Seq | WRKY |
| 435 | WRKY65 | 20576 | Plant Cistrome Database | DAP-Seq | WRKY |
| 436 | WRKY6 | 5323 | Plant Cistrome Database | DAP-Seq | WRKY |
| 437 | WRKY70 | 8244 | Plant Cistrome Database | DAP-Seq | WRKY |
| 438 | WRKY71 | 9533 | Plant Cistrome Database | DAP-Seq | WRKY |
| 439 | WRKY75 | 29735 | Plant Cistrome Database | DAP-Seq | WRKY |
| 440 | WRKY7 | 1926 | Plant Cistrome Database | DAP-Seq | WRKY |
| 441 | WRKY8 | 23621 | Plant Cistrome Database | DAP-Seq | WRKY |
Sheet 4: Information of prime motif for every TF of Dataset 'A'
| S.No. | TF | Motif | Coverage (%) |
|---|---|---|---|
| 1 | ABF1 | ACACGTGGCAA | 96.52443685 |
| 2 | ABF3 | CCACGTGTCTT | 96.12644528 |
| 3 | ABF4 | CCACGTGTCAT | 96.45850994 |
| 4 | ABI5 | TTGCCACGTGTC | 87.24088635 |
| 5 | ANAC032 | ACGCGTCGT | 89.218 |
| 6 | ANAC102 | CGACACGTC | 91.4415 |
| 7 | AP1 | GGGGTCCTA | 91.2963 |
| 8 | AZF1 | TTGATGGGGTT | 89.69266151 |
| 9 | BZIP28 | AAGCCACGGGT | 93.01270417 |
| 10 | CCA1 | CCGCCCCGG | 84.2454 |
| 11 | CRY2 | CGACGACGGAA | 93.26710817 |
| 12 | DELLA | CGCGTAAAA | 88.9697 |
| 13 | DREB2A | TCGGTTATAT | 96.10218594 |
| 14 | FBH3 | ACGTGGGCC | 88.1206 |
| 15 | FHY1 | CGGGATCCCTT | 97.22470904 |
| 16 | FHY3 | AAACGCGTCAT | 95.94594595 |
| 17 | FIE | CCCCAACCC | 76.4222 |
| 18 | FLM | TAGGACCCC | 82.4163 |
| 19 | GBF2 | TTGACACGTGG | 95.21389514 |
| 20 | HAT22 | TACGTGGCC | 85.5933 |
| 21 | HB5 | TATGGGCCC | 87.3902 |
| 22 | HB7 | GTGGGCCCA | 81.746 |
| 23 | HBI | TCCGGGGGC | 87.5 |
| 24 | HSFA1A | TTCGCCGCCGA | 96.22770919 |
| 25 | HSFA6A | ACGTGGGCC | 81.479 |
| 26 | IBH1 | GGCCGAATCAA | 96.52777778 |
| 27 | JAG | TTCACGTACGT | 91.40142096 |
| 28 | KAN1 | TTATATATATT | 91.51017144 |
| 29 | LEC1 | CCACGTGGCAA | 93.95186049 |
| 30 | LFY | TACGGCCCA | 83.472 |
| 31 | MYB3 | ACGTGGGCC | 86.1625 |
| 32 | NFYB2 | AAGACACGTGG | 92.09588682 |
| 33 | NFYC2 | AAGACACGTGG | 95.50911924 |
| 34 | PIF1 | CCACGTGTCAT | 94.73684211 |
| 35 | PIF4 | CCCACGTGTTT | 94.61862286 |
| 36 | PIF5 | TCACGTGGGCT | 96.32918887 |
| 37 | REV | TGGGCCCAT | 90.514 |
| 38 | RGA | CGTGGGCCC | 79.0311 |
| 39 | SOC1 | TTTTGGGCCCA | 93.66476023 |
| 40 | TOC1 | GCCGTCCGC | 93.19371728 |
| 41 | WRKY33 | TTATATTATAA | 91.82324949 |
| 42 | ZAT6 | GGCCCACGT | 82.9334 |
| 43 | AP2 | CGGACGCCC | 87.84194529 |
| 44 | AT5G04760 | AGACGTGGC | 91.61023078 |
| 45 | BBM | CCACGAGGA | 89.96696227 |
| 46 | ERF115 | GGCACCGAA | 95.7147906 |
| 47 | GBF3 | TCCACGTGG | 94.12760154 |
| 48 | HB6 | ACACGTCGC | 88.5862129 |
| 49 | MYB3R3 | AAAGGCCGC | 92.02898551 |
| 50 | MYB44 | TCCACGTGG | 90.73768833 |
| 51 | PhyA | ACCCGGCCT | 97.17868339 |
| 52 | RD26 | ACGTCGCCA | 90.70929623 |
| 53 | SEP3 | GTGGGCCAA | 90.55165496 |
| 54 | SVP | TCCAAGCTG | 75.95842956 |
Sheet 5: Information of prime motif for every TF of Dataset 'B'
| S.No. | TF | Motif | Coverage (%) |
|---|---|---|---|
| 1 | ABF2 | GGACACGTG | 95.43279687 |
| 2 | ABI5 | CACGTGACC | 95.78685151 |
| 3 | ABR1 | AATAGCCGC | 87.38345736 |
| 4 | Adof1 | GCGCCTTT | 90.2589 |
| 5 | AGL15 | GGGGAAAG | 87.9929 |
| 6 | AGL63 | CTGGCACC | 82.9152 |
| 7 | AIL7 | TCTCGGGAA | 86.22347949 |
| 8 | ANAC004 | AGCAGGCTT | 80.69822088 |
| 9 | ANAC005 | TGCAGGAAG | 78.45131013 |
| 10 | ANAC013 | TGCAGCAAG | 82.32938719 |
| 11 | ANAC016 | GCCAGGAAA | 80.76849551 |
| 12 | ANAC017 | TCCGAGGAA | 83.09707632 |
| 13 | ANAC020 | CTTGCTGCA | 80.38198836 |
| 14 | ANAC028 | GAAGCAGCT | 81.28905771 |
| 15 | ANAC034 | CGTTACCG | 94.8944 |
| 16 | ANAC038 | ACGTGACGT | 81.39572539 |
| 17 | ANAC042 | CATCGGAGA | 84.68085106 |
| 18 | ANAC045 | AGCTTCCGT | 82.72417834 |
| 19 | ANAC046 | ACGTACGC | 92.2997 |
| 20 | ANAC047 | CACGTGGC | 89.4942 |
| 21 | ANAC050 | CTTGCTGCA | 81.37823688 |
| 22 | ANAC053 | CTTGCTGCA | 82.66646304 |
| 23 | ANAC055 | CACGTGGC | 89.6313 |
| 24 | ANAC057 | ACGCAAGCT | 85.62475519 |
| 25 | ANAC058 | GCCACGTG | 91.5113 |
| 26 | ANAC062 | GGCGAAGC | 88.7073 |
| 27 | ANAC070 | GCTTGCGC | 88.9159 |
| 28 | ANAC071 | CGCCAAGG | 88.1534 |
| 29 | ANAC075 | AGCTTCCTG | 86.0109539 |
| 30 | ANAC079 | ACGTCACGT | 76.81931132 |
| 31 | ANAC083 | ACGTACGC | 93.2138 |
| 32 | ANAC087 | AACGTCACG | 84.37255821 |
| 33 | ANAC092 | CGCGTTAC | 95.0019 |
| 34 | ANAC094 | ACGTCAACG | 89.56639566 |
| 35 | ANAC096 | GCGCAAGC | 89.5171 |
| 36 | ANAC103 | ACTTGCTGC | 83.5784877 |
| 37 | ANL2 | CCGAGTCT | 79.29366231 |
| 38 | AREB3 | GTCACGTGA | 96.4479099 |
| 39 | ARF2 | GCCGGCAC | 79.32943223 |
| 40 | ASL18 | CCAGGCGT | 96.32474477 |
| 41 | AT1G01250 | ATCGACGGA | 86.89655172 |
| 42 | AT1G12630 | TTCTGCCGA | 84.73999231 |
| 43 | At1g19000 | GGATCCGC | 88.7445 |
| 44 | AT1G19040 | CGTGAAGCA | 84.30583501 |
| 45 | At1g19210 | ATCGACGGT | 87.92035989 |
| 46 | AT1G20910 | TGGCTCCA | 79.26165313 |
| 47 | At1g22810 | CTGCCCAC | 96.95481336 |
| 48 | AT1G24250 | TGGCAGCTA | 75.23510972 |
| 49 | At1g25550 | GAGGATCCT | 89.33483952 |
| 50 | AT1G28160 | CTACCGGCG | 92 |
| 51 | At1g36060 | ACCGTCGAT | 90.01239157 |
| 52 | AT1G44830 | CACCGGTGA | 88.74617737 |
| 53 | AT1G47655 | CCTTTCGCT | 75.21410579 |
| 54 | At1g49010 | GGATCCGC | 86.9606 |
| 55 | AT1G49560 | GAGGATCCT | 90.03146453 |
| 56 | At1g64620 | GGGAAAAGT | 77.4565454 |
| 57 | At1g68670 | AGAATCCCC | 86.74203494 |
| 58 | AT1G69570 | ACTTTTTCC | 82.25548612 |
| 59 | At1g69690 | ATGGGCCCA | 92.96482412 |
| 60 | AT1G71450 | CTTACGCCG | 81.01842124 |
| 61 | At1g72010 | TGGGTCCCA | 93.49329875 |
| 62 | AT1G72740 | CCCTAGGG | 90.29 |
| 63 | At1g74840 | CTGGATCCA | 80.00786705 |
| 64 | AT1G76880 | GGCGGTAAA | 79.02240326 |
| 65 | AT1G77200 | ATCGACGGT | 84.75126305 |
| 66 | At1g78700 | CTCACGTGT | 98.29026617 |
| 67 | At2g01060 | ATTCCCGG | 94.1155 |
| 68 | At2g03500 | GGGGATCC | 88.8775 |
| 69 | AT2G15740 | AATCGACGC | 95.85508136 |
| 70 | AT2G20110 | TCGTGGCC | 75.76894224 |
| 71 | AT2G20400 | GGGGATCC | 87.2076 |
| 72 | AT2G28810 | GCCTTTGC | 89.4585 |
| 73 | AT2G33550 | AAGGCCCTT | 86.7121545 |
| 74 | At2g33710 | TTAGCGGCG | 89.35885065 |
| 75 | AT2G40260 | GAGGATTCT | 85.92808907 |
| 76 | At2g44940 | TGACGGACA | 86.77125702 |
| 77 | At2g45680 | GTATTTAA | 75.45145718 |
| 78 | At3g04030 | GAGGCTCC | 87.2661 |
| 79 | At3g09600 | TTAAATATT | 97.01649226 |
| 80 | AT3G09735 | TCCTCGAGA | 88.47531338 |
| 81 | AT3G10030 | ACGTTACCG | 93.85964912 |
| 82 | AT3G10113 | TAAATATTT | 97.32100261 |
| 83 | AT3G10580 | CCGGACCT | 84.3629 |
| 84 | At3g11280 | GGATCCGC | 87.4938 |
| 85 | AT3G12130 | AGCCTTTGC | 77.15215622 |
| 86 | At3g12730 | GGATCCGC | 88.9023 |
| 87 | At3g14180 | TCGGCGAAA | 96.79661529 |
| 88 | AT3G16280 | ACTGACGGT | 81.67067308 |
| 89 | At3g24120 | ATTCCCGG | 93.9544 |
| 90 | AT3G25990 | ACCCGGGT | 84.34 |
| 91 | AT3G42860 | ACGTCGACT | 93.99585921 |
| 92 | At3g45610 | CAGAAAGGT | 82.19787843 |
| 93 | AT3G52440 | GGGAAAAGT | 78.18815102 |
| 94 | AT3G60490 | ACTGACGGT | 82.22222222 |
| 95 | At3g60580 | TGCACTAGC | 81.52733845 |
| 96 | AT4G00250 | ATCCGGATC | 82.4781191 |
| 97 | At4g01280 | ATATATTAA | 96.04130107 |
| 98 | At4g16750 | TCTGTCGAC | 85.22038655 |
| 99 | AT4G18450 | CACCGATGC | 85.20499109 |
| 100 | At4g18890 | CACGTGCCA | 97.4566474 |
| 101 | AT4G26030 | TGCAGCTAC | 79.03667698 |
| 102 | At4g31060 | CGGCGGTTA | 81.99361192 |
| 103 | At4g32800 | TGACGGACA | 84.81079651 |
| 104 | At4g36780 | CGACACGTG | 97.16840537 |
| 105 | AT4G37180 | AGGATCCGA | 82.97256098 |
| 106 | At4g38000 | AGCCTTTC | 90.4899 |
| 107 | AT5G02460 | GCCTTTCC | 86.1828 |
| 108 | At5g04390 | ACTCACTGG | 85.90604027 |
| 109 | AT5G05550 | ATCTCCGGC | 96.49622711 |
| 110 | At5g05790 | GATCCGGAT | 85.40915395 |
| 111 | At5g08330 | TGGGTCCCA | 93.10463122 |
| 112 | At5g08520 | GGATCCGC | 85.8047 |
| 113 | At5g08750 | GGCGGTAAA | 84.7107438 |
| 114 | At5g18450 | GCGCCGGTT | 91.76715177 |
| 115 | AT5G22990 | TCGAGTTCG | 94.1404655 |
| 116 | AT5G23930 | GAGCCGGCT | 92.16357413 |
| 117 | At5g29000 | GGGGATCC | 86.9685 |
| 118 | AT5G45580 | GCGGATCC | 88.6798 |
| 119 | At5g47390 | ATCCGGATC | 75.47094188 |
| 120 | AT5G47660 | GTTACCGG | 85.4929 |
| 121 | At5g52660 | TAAATATTT | 97.2968822 |
| 122 | AT5G56840 | CGATATCCA | 80.54007579 |
| 123 | At5g58900 | GGATCCGG | 84.9871 |
| 124 | AT5G60130 | TTAGAAATT | 93.57269968 |
| 125 | AT5G61620 | GGATCCGC | 86.6439 |
| 126 | At5g62940 | TTTTAAGAA | 91.95810047 |
| 127 | AT5G63260 | GCCGCTTT | 91.5774 |
| 128 | At5g65130 | CGCCGGTAA | 92.26907631 |
| 129 | At5g66730 | TGACGTCAA | 79.52518363 |
| 130 | AT5G66940 | GCCCTTTC | 86.6292 |
| 131 | ATAF1 | ACGTCACGT | 81.70055453 |
| 132 | AtGRF6 | GCCGTCGG | 96.44543659 |
| 133 | ATHB13 | TATAATTAA | 96.82765152 |
| 134 | ATHB15 | TGATCATCA | 93.1936372 |
| 135 | ATHB18 | CCCATGATT | 92.8997391 |
| 136 | ATHB20 | TAATATTTA | 97.42852309 |
| 137 | ATHB21 | AATAATTTA | 97.70540675 |
| 138 | ATHB23 | CCACGTGG | 79.16666667 |
| 139 | ATHB24 | GGGTCCAC | 79.02240326 |
| 140 | ATHB25 | CCCGAGAG | 75.83892617 |
| 141 | AtHB32 | GCTGGCCC | 75.45145718 |
| 142 | ATHB33 | TTAATCGCG | 79.26165313 |
| 143 | ATHB34 | ATTTAATAA | 97.88299567 |
| 144 | ATHB40 | GCCATCATT | 83.5380117 |
| 145 | ATHB5 | GCCATCATT | 85.81280576 |
| 146 | ATHB53 | AATAATTTA | 97.87075725 |
| 147 | ATHB6 | GGCCATGAT | 81.32966249 |
| 148 | ATHB7 | AATCATGGG | 91.20195931 |
| 149 | AtIDD11 | ACGACGAC | 84.8827 |
| 150 | ATY13 | AGCTGCTAC | 76.53311529 |
| 151 | BAM8 | CGCACGTGT | 97.90804097 |
| 152 | BBX31 | TCTTCAAAA | 88.87630402 |
| 153 | bHLH10 | TGACGGACA | 85.23622047 |
| 154 | bHLH122 | CAACTCGCT | 89.53233831 |
| 155 | bHLH28 | ACACGTACG | 85.8506457 |
| 156 | bHLH31 | CACGTGGGT | 95.84472389 |
| 157 | bHLH80 | AGCTACTGG | 81.34815599 |
| 158 | BIM1 | CACGTGGGC | 95.51777434 |
| 159 | BIM2 | CCACCAGC | 96.82765152 |
| 160 | BOS1 | GGCCCAAC | 89.6092 |
| 161 | BPC1 | GCTCTGGC | 87.5417 |
| 162 | bZIP16 | TCACGTGAC | 95.18086625 |
| 163 | bZIP18 | ACCAGCTGT | 91.9737862 |
| 164 | bZIP28 | CGTCACGTG | 95.07531866 |
| 165 | bZIP3 | GCTCACGTG | 88.5111513 |
| 166 | bZIP44 | AGCACGTGA | 87.86860769 |
| 167 | bZIP48 | GCGACGTGA | 92.41919687 |
| 168 | bZIP50 | TGACGTCAG | 92.07898423 |
| 169 | bZIP52 | ACCAGCTGT | 90.56660821 |
| 170 | bZIP53 | TCACGTGAC | 92.60002293 |
| 171 | bZIP68 | GTCACGTGA | 93.79699248 |
| 172 | BZR1 | CACGTGTCG | 96.95481336 |
| 173 | CAMTA1 | CAACGCGTT | 95.31700288 |
| 174 | CAMTA5 | AGAACGCGT | 91.72056921 |
| 175 | CBF1 | CTGTCGACA | 88.01639521 |
| 176 | CBF2 | TGTCGACGT | 85.62776115 |
| 177 | CBF3 | ACGTCGACA | 86.4835978 |
| 178 | CBF4 | TCGTCGACA | 87.08515651 |
| 179 | CDF3 | TGCCCTTTT | 77.94478528 |
| 180 | CDM1 | TCCGGAGAT | 86.204731 |
| 181 | CEJ1 | TTGACCGTC | 84.947726 |
| 182 | COG1 | AGGCCTTTT | 77.69948917 |
| 183 | CRC | CACCATGG | 94.5958 |
| 184 | CRF10 | CAACCGGGG | 81.93447738 |
| 185 | CRF4 | AAGCGCCGG | 94.92344883 |
| 186 | CUC1 | CGCAAGCG | 90.7366 |
| 187 | CUC2 | CGCGTTAC | 95.1363 |
| 188 | CUC3 | CGTACCGT | 91.0511 |
| 189 | DAG2 | CGTAAAGGT | 80.59002383 |
| 190 | DDF1 | TGTCGACGT | 96.15866388 |
| 191 | DDF2 | ACGTCTGCA | 87.11656442 |
| 192 | DEAR2 | AACCGTCGA | 85.98846717 |
| 193 | DEAR3 | ATCGACGGT | 85.81761006 |
| 194 | DEAR5 | TGTCAGCGA | 88.69832894 |
| 195 | DEL2 | AAATTATA | 79.40797941 |
| 196 | dof24 | CCCCTTTT | 88.4655 |
| 197 | dof42 | AGAAAAGCC | 79.65043695 |
| 198 | dof43 | CACCTTTCT | 81.83009963 |
| 199 | dof45 | GCCGCTTT | 87.2531 |
| 200 | DREB19 | TGACCGTCA | 81.11332008 |
| 201 | E2FA | TTGGCGCCA | 83.82620883 |
| 202 | EIN3 | ATGCAGCGA | 79.23904053 |
| 203 | EPR1 | ATAATTATA | 94.48836727 |
| 204 | ERF10 | GTCGCCGAA | 94.03450426 |
| 205 | ERF105 | CGCCGGCAA | 92.35815243 |
| 206 | ERF11 | CCGCAGTTG | 88.54275287 |
| 207 | ERF115 | TTCAGCGGC | 92.78572019 |
| 208 | ERF13 | AAAGTCGGC | 88.22729175 |
| 209 | ERF15 | AAGCCGGCT | 89.031735 |
| 210 | ERF2 | ACCGACGGC | 96.14325069 |
| 211 | ERF3 | CCGGCGACT | 94.45950015 |
| 212 | ERF38 | TCACTGTCG | 85.73758608 |
| 213 | ERF4 | CGGCGAATT | 89.48199829 |
| 214 | ERF5 | ACTGCGGCG | 93.97163121 |
| 215 | ERF7 | GGCGGCAAT | 88.60630722 |
| 216 | ERF8 | ATATCGCCG | 90.98137033 |
| 217 | ESE1 | CGCAGCCGA | 93.71693122 |
| 218 | ESE3 | TAACCGGCG | 93.06389365 |
| 219 | FAR1 | CAACGCGTG | 96.0292581 |
| 220 | FRS9 | ACGGGTTG | 79.03667698 |
| 221 | FUS3 | GGGCATGCA | 88.5486834 |
| 222 | GATA1 | CATCGACGT | 87.21649485 |
| 223 | GATA11 | AGATCCGGA | 92.75960622 |
| 224 | GATA19 | ACCGATCGG | 96.03960396 |
| 225 | GATA20 | CTCCCTGC | 96.79661529 |
| 226 | GATA4 | TGGATCTCC | 93.66852886 |
| 227 | GBF3 | TCACGTGAC | 93.11463141 |
| 228 | GBF5 | TCGACGTGA | 90.20532319 |
| 229 | GBF6 | GCTCACGTG | 85.13240348 |
| 230 | GRF9 | TCTGCCAGA | 95.87194608 |
| 231 | GT1 | TGGTTAGCC | 87.86127168 |
| 232 | GT2 | ACGGTAACT | 79.40797941 |
| 233 | GT3a | ACCGCGTG | 83.9909 |
| 234 | GTL1 | AGTTACCGT | 78.67128423 |
| 235 | HAT2 | CGGGGTCT | 75.47094188 |
| 236 | HDG1 | ATAATTATA | 96.44543659 |
| 237 | HSF21 | CCTCGAGAA | 95.76719577 |
| 238 | HSF3 | TCCTCGAGA | 90.20353901 |
| 239 | HSF6 | AGAACCTCC | 94.29502852 |
| 240 | HSF7 | CTCCTGGAA | 91.2606169 |
| 241 | HSFA6B | TCCTCGAGA | 88.49281338 |
| 242 | HSFB3 | TTCCGGAAG | 85.99487617 |
| 243 | HSFC1 | CTGCAGGAA | 80.45247785 |
| 244 | HY5 | TGACGTCTC | 90.84812623 |
| 245 | IDD4 | TGTCGGCA | 88.5541 |
| 246 | IDD5 | CTGTCGCC | 81.2521 |
| 247 | IDD7 | CTTGGCCG | 83.0085 |
| 248 | JKD | TGTCGGCA | 89.2593 |
| 249 | LBD13 | TTTCCGGCA | 88.77523553 |
| 250 | LBD18 | TGCCGGAAA | 89.35057244 |
| 251 | LBD19 | TGCCGGAAA | 87.7674438 |
| 252 | LBD2 | TCGCCGAAA | 88.50045241 |
| 253 | LBD23 | TTTCGCGGA | 86.56099242 |
| 254 | LCL1 | TCATATATA | 91.00358423 |
| 255 | LEP | GCCGCGAAT | 91.20879121 |
| 256 | LHY1 | TTAAATATT | 97.620043 |
| 257 | LMI1 | TAATATTTA | 97.27593336 |
| 258 | LOB | CTCCGCAGA | 92.60780287 |
| 259 | MGP | CTTGGCCG | 84.0095 |
| 260 | MP | GTGTCGGC | 89.6658 |
| 261 | MS188 | ACCTAGGTG | 78.30337802 |
| 262 | MYB101 | ACCGGTTAC | 80.45375218 |
| 263 | MYB107 | GGTGCACC | 92.334 |
| 264 | MYB116 | ACCTAGGCA | 77.91541123 |
| 265 | MYB118 | CGTTACGGT | 85.06040892 |
| 266 | MYB119 | CGTTACGGA | 85.18351867 |
| 267 | MYB121 | TAGGTACCG | 79.29366231 |
| 268 | MYB13 | AGCTACCGA | 77.53384559 |
| 269 | MYB27 | ACCTGAGG | 93.3636 |
| 270 | MYB33 | CCGGTTGC | 86.8476 |
| 271 | MYB3R1 | AACCGGTAC | 76.16380662 |
| 272 | MYB3R4 | AACCGGTGA | 86.15776081 |
| 273 | MYB3R5 | ACCGGTGAA | 83.77679231 |
| 274 | MYB44 | ACGGTTACC | 81.50862628 |
| 275 | MYB49 | GGTGACCC | 90.8142 |
| 276 | MYB55 | TGCAGCTAC | 82.46940603 |
| 277 | MYB56 | CGGTAACGA | 81.47044213 |
| 278 | MYB57 | GTTCGGCC | 86.7763 |
| 279 | MYB58 | GGTGCACCA | 75.07142199 |
| 280 | MYB61 | TGCAGCTAC | 82.34962829 |
| 281 | MYB62 | GTTCGGCC | 91.1681 |
| 282 | MYB63 | TGGTGCCAC | 75.76894224 |
| 283 | MYB65 | TGGTAACCG | 82.20720721 |
| 284 | MYB67 | CAGCTAGCT | 75.83892617 |
| 285 | MYB70 | GGTAACGGT | 78.87409201 |
| 286 | MYB73 | GGTAACGGT | 77.8358209 |
| 287 | MYB74 | CAGCTAGGT | 82.58738974 |
| 288 | MYB77 | ACGGTTACC | 79.40432087 |
| 289 | MYB81 | TGGTAACCG | 83.00835655 |
| 290 | MYB83 | TGCAGCTAC | 83.14715212 |
| 291 | MYB88 | AAGAGGCTC | 90.6336817 |
| 292 | MYB93 | CAGCTAGCT | 77.48624088 |
| 293 | MYB96 | CCAGCTGG | 93.2868 |
| 294 | MYB98 | CGTTACGGA | 88.61101116 |
| 295 | MYB99 | GGTGCACC | 90.3202 |
| 296 | NAC2 | CTTCCTGCA | 78.21991298 |
| 297 | NAM | ACGTCACGT | 81.06670127 |
| 298 | NAP | CGGTACGT | 93.3164 |
| 299 | NGA4 | ACCAGCTGA | 87.56152125 |
| 300 | NLP7 | AGAGGCCTT | 81.43021915 |
| 301 | NST1 | GCGCAAGC | 88.3973 |
| 302 | NTL8 | CAGGAAGCT | 84.37341451 |
| 303 | NTM1 | GCGCAAGG | 86.083 |
| 304 | NTM2 | AAGCAGCCT | 77.97001153 |
| 305 | OBP1 | CGCCCTTT | 88.456 |
| 306 | OBP3 | CTTTACAGA | 82.49630724 |
| 307 | OBP4 | AGCACCTTT | 83.16007573 |
| 308 | PDF2 | GGAGATCCT | 79.16666667 |
| 309 | PHV | ATATTTAAT | 96.32474477 |
| 310 | PLT3 | TCCCTGGAA | 84.2883549 |
| 311 | PUCHI | AAACCGACG | 91.93608324 |
| 312 | RAP21 | CATCGACGG | 89.53875416 |
| 313 | Rap210 | TGTCGGCAA | 91.6984127 |
| 314 | RAP211 | GCCGGCAAA | 93.05606901 |
| 315 | RAP212 | CGGCGAATT | 89.60584386 |
| 316 | RAP26 | TTAGCGGCG | 90.14232385 |
| 317 | RAV1 | GCTGCTGC | 80.6988 |
| 318 | REM19 | CCACGTGG | 76.81931132 |
| 319 | RKD2 | TTCCGGGA | 94.5158 |
| 320 | RRTF1 | GCGGCAATT | 91.16585791 |
| 321 | RVE1 | CGGATATCT | 83.05122997 |
| 322 | SGR5 | TTGTCTGTG | 79.32943223 |
| 323 | SMB | GCTTGCGC | 89.3893 |
| 324 | SND2 | ACGCTGCTT | 77.89640504 |
| 325 | SND3 | TGCTGCTG | 92.1429 |
| 326 | SOL1 | CCACGTGG | 76.53311529 |
| 327 | SPL1 | GGACGTACA | 87.56568779 |
| 328 | SPL13 | TCGTACGTG | 85.072095 |
| 329 | SPL14 | GTACGTACG | 93.98971926 |
| 330 | SPL15 | CGTACAGGT | 82.43331165 |
| 331 | SPL5 | CACGTACGA | 83.65359885 |
| 332 | SPL9 | ACGTACAGG | 82.63938914 |
| 333 | SRS7 | CCTAGGCTT | 85.03089144 |
| 334 | STOP1 | CCTGGTGAA | 85.84070796 |
| 335 | STZ | AGTCACTGC | 88.12314295 |
| 336 | SVP | CTGGGCCA | 88.6934 |
| 337 | TBP3 | CCCTAGGG | 87.9206 |
| 338 | TCP20 | GGGTCCGAC | 79.86289632 |
| 339 | TCP24 | TGGTCCTCA | 89.91646778 |
| 340 | TCP3 | TGGACCCTA | 86.5819209 |
| 341 | TCP7 | TGGACCCCA | 88.1237525 |
| 342 | TCX2 | TCGTGGCC | 76.16380662 |
| 343 | TGA1 | TGACGTCAC | 95.40276319 |
| 344 | TGA10 | TGACGTCGT | 93.76363636 |
| 345 | TGA2 | CTGACGTCA | 90.35992042 |
| 346 | TGA3 | CTGACGTCA | 95.39263981 |
| 347 | TGA4 | TGACGTCGT | 95.28402367 |
| 348 | TGA5 | GTGACGTCA | 96.00614439 |
| 349 | TGA6 | TGACGTCAC | 94.98478092 |
| 350 | TGA9 | TGACGTCAC | 94.77967554 |
| 351 | TINY | TGTCGGACA | 85.35211268 |
| 352 | TRP1 | AAACTTTA | 79.23904053 |
| 353 | TRP2 | TTTAAACT | 79.40432087 |
| 354 | VIP1 | TCCAGCTGT | 92.0551543 |
| 355 | VND1 | ACGGAAGCT | 80.36612633 |
| 356 | VND2 | ACGCTACG | 93.9193 |
| 357 | VND3 | ACGCTGCTT | 77.74716866 |
| 358 | VND4 | GCTTGCGC | 89.6062 |
| 359 | VND6 | CTGCTGCT | 90.6535 |
| 360 | VRN1 | AGGCCTAT | 75.3415 |
| 361 | WIP5 | CCTCGAGG | 92.0677 |
| 362 | WRKY14 | AACGTCGAC | 88.10891458 |
| 363 | WRKY15 | GGTCGACC | 93.1993 |
| 364 | WRKY18 | TTGACGACG | 81.21844164 |
| 365 | WRKY20 | AGTCGACGA | 86.7148667 |
| 366 | WRKY21 | GTCGACGAT | 85.75146389 |
| 367 | WRKY22 | GGTCGACC | 91.3985 |
| 368 | WRKY24 | GGTCGACCT | 75.05565568 |
| 369 | WRKY25 | GGTCGACC | 93.5698 |
| 370 | WRKY27 | GGTCGACC | 93.2413 |
| 371 | WRKY28 | GGTCGACC | 89.9119 |
| 372 | WRKY29 | AACGTCGAC | 85.28536508 |
| 373 | WRKY3 | GTCGACTGT | 87.33488733 |
| 374 | WRKY30 | GAGTCGACT | 91.31229995 |
| 375 | WRKY31 | CGTCGACTT | 87.87618229 |
| 376 | WRKY33 | TGGTCGACC | 75.45145718 |
| 377 | WRKY40 | GGTCGACC | 91.3832 |
| 378 | WRKY45 | GGTCGACC | 90.1343 |
| 379 | WRKY50 | GTCGACGA | 92.7586 |
| 380 | WRKY55 | CAGTCGACT | 86.6504317 |
| 381 | WRKY6 | CCGAGGGA | 96.15866388 |
| 382 | WRKY65 | AACGTCGAC | 87.36391913 |
| 383 | WRKY7 | GGTCGACCT | 76.9470405 |
| 384 | WRKY70 | AGGTCGACT | 88.3915575 |
| 385 | WRKY71 | GGTCGACC | 91.5032 |
| 386 | WRKY75 | GGTCGACC | 93.2907 |
| 387 | WRKY8 | GGTCGACC | 90.6482 |
Sheet 6: Information of experimentally derived motif from JASPAR database on ChIP and DAP-seq TFs
| S.No. | TF | Total peaks | Covered peaks | Coverage (%) | Consensus | Rank | p-value |
|---|---|---|---|---|---|---|---|
| 1 | AGL15 | 2257 | 453 | 20.0709 | CTTTCCTTTTTTGGAAAGT | 1 | 0.00240911 |
| 2 | BZR1 | 1018 | 80 | 7.85855 | CCACACGTGCGACTTTTTTT | 1 | 0.000918004 |
| 3 | HY5 | 10140 | 1487 | 14.6647 | AATGCCACGTGGCATT | 7 | 0.00085681 |
| 4 | SVP | 20687 | 1281 | 6.19229 | ATTACCAAAAAAGGAAAGAA | 3 | 0.000254927 |
| 5 | FUS3 | 3266 | 2782 | 85.1806 | AAACATGCAT | 1 | 3.43E-03 |
| 6 | MYB77 | 27263 | 6842 | 25.0963 | AAGTGACAGTTAC | 1 | 0.000425009 |
| 7 | RAV1 | 14740 | 7972 | 54.0841 | CAGCAACAGAAA | 4 | 2.81E-09 |
| 8 | RAV1 | 14740 | 4199 | 28.4871 | ATCACCTGAGGC | 3 | 0.00252729 |
| 9 | SPL14 | 2529 | 2239 | 88.533 | TCCGTACAATT | 2 | 0.00301584 |
| 10 | TGA1 | 12377 | 10976 | 88.6806 | TGGTGACGTAA | 1 | 2.99E-05 |
| 11 | ABI5 | 9233 | 7306 | 79.1292 | TGACACGTGG | 1 | 1.82E-03 |
| 12 | ABF2 | 2299 | 1128 | 49.0648 | CATGACACGTGTT | 1 | 3.10E-04 |
| 13 | BIM1 | 647 | 569 | 87.9444 | AGTCACGTGACT | 1 | 0.00263206 |
| 14 | BIM2 | 40836 | 4854 | 11.8866 | AAAGGCACGTGCCTTT | 8 | 2.19E-04 |
| 15 | bZIP68 | 9044 | 2877 | 31.8111 | TGCCACGTCAGCATC | 3 | 1.44E-05 |
| 16 | CDF3 | 22820 | 15207 | 66.6389 | ATAAAAAGTGAA | 1 | 0.000359122 |
| 17 | CRF4 | 2482 | 1801 | 72.5624 | CCGCCGCCACCG | 1 | 0.00289419 |
| 18 | ERF4 | 15193 | 9212 | 60.6332 | GCCGCCGTCATA | 1 | 0.000789394 |
| 19 | ERF7 | 17694 | 11671 | 65.9602 | CGCCGCCATT | 1 | 6.91E-05 |
| 20 | ERF8 | 12829 | 3061 | 23.86 | CGGTGACGGCGGCGGTGG | 1 | 0.00165542 |
| 21 | ERF105 | 14787 | 5891 | 39.839 | GTGGCGGCGGCGGAG | 1 | 0.000203768 |
| 22 | ERF11 | 7672 | 4719 | 61.5094 | CCGCCGCCACCG | 3 | 5.72E-04 |
| 23 | ERF13 | 9556 | 9220 | 96.4839 | CGCCGCCA | 3 | 0.00308845 |
| 24 | ERF3 | 3321 | 2926 | 88.106 | GCCGCCGCCGC | 1 | 0.00193623 |
| 25 | GATA11 | 3149 | 3112 | 98.825 | TAGATCTG | 1 | 0.00201444 |
| 26 | MYB55 | 10051 | 9535 | 94.8662 | ACCTACCG | 8 | 0.00287828 |
| 27 | TGA5 | 2604 | 1695 | 65.0922 | ATGATGACGTCATCA | 1 | 0.00332933 |
| 28 | SPL1 | 12940 | 4811 | 37.1793 | ATTGTACGGATAA | 3 | 0.00322949 |
| 29 | SPL5 | 27217 | 15503 | 56.9607 | AATTGTACGGAC | 1 | 0.00043355 |
| 30 | TCP20 | 1167 | 227 | 19.4516 | AAAAGTGGGACCCACATTT | 1 | 1.50E-03 |
| 31 | TGA2 | 16587 | 6479 | 39.0607 | ATGATGACGTCATCA | 1 | 2.92E-05 |
| 32 | TGA6 | 28254 | 13597 | 48.1242 | ATGATGACGTCATCA | 1 | 0.000123057 |
| 33 | WRKY15 | 29394 | 6053 | 20.5926 | AAAAAAGTCAACGGTT | 7 | 1.47E-04 |
| 34 | WRKY18 | 30301 | 17327 | 57.1829 | ATGGTCAACG | 6 | 4.83E-03 |
| 35 | WRKY21 | 1537 | 160 | 10.4099 | AAAAAAAGTCAACGCAAAAA | 1 | 0.0023834 |
| 36 | WRKY25 | 20979 | 3468 | 16.5308 | AAATTGGTCAACGATA | 4 | 0.000242448 |
| 37 | WRKY30 | 4374 | 2752 | 62.9172 | AAAGTCAACGCTTA | 5 | 0.000305396 |
| 38 | WRKY40 | 7114 | 5425 | 76.2581 | AAAGTCAAAA | 7 | 8.56E-09 |
| 39 | WRKY45 | 13329 | 5908 | 44.3244 | AAAAAAGTCAACGAT | 7 | 9.67E-04 |
| 40 | WRKY75 | 29735 | 29409 | 98.9036 | AAGTCAAC | 9 | 8.88E-05 |
| 41 | WRKY8 | 23621 | 6083 | 25.7525 | AAAAAAGTCAACGATT | 6 | 0.000409345 |
| 42 | JKD | 2160 | 221 | 10.2315 | TTTTTTTTTGTCGTTTTCTG | 3 | 0.00446514 |
| 43 | MGP | 3771 | 346 | 9.17529 | CAGAAAACGACAAAAAAAAA | 4 | 0.00239986 |
| 44 | SGR5 | 18969 | 8459 | 44.5938 | AAAAAAAGACAAAAA | 5 | 0.00494099 |
| 45 | TCX2 | 24443 | 11442 | 46.8109 | ATTTAAAATTCAAAT | 8 | 0.00342864 |
| 46 | MYB118 | 4304 | 1639 | 38.0809 | AAAATAACCGTTACAAA | 3 | 1.43E-03 |
| 47 | MYB3R5 | 2706 | 981 | 36.2528 | AAAATATGACCGTTG | 6 | 2.19E-04 |
| 48 | MYB101 | 10887 | 3150 | 28.9336 | AACCGTAACCGAATT | 1 | 0.00283513 |
| 49 | MYB56 | 6039 | 2613 | 43.2688 | ATAAATAACGGTATT | 4 | 8.74E-05 |
| 50 | MYB81 | 3231 | 2751 | 85.1439 | GTAACCGAATT | 1 | 0.000430339 |
| 51 | MYB119 | 15557 | 13714 | 88.1532 | TAACCGTTACA | 1 | 0.000908697 |
| 52 | MYB65 | 12876 | 3594 | 27.9124 | ACCGTAACCGTAA | 1 | 0.00239262 |
| 53 | MYB3R1 | 22319 | 7120 | 31.9011 | ATCTAACGGTCAA | 6 | 0.00175337 |
| 54 | MYB3R4 | 1965 | 823 | 41.883 | AAATATAACCGTTGG | 3 | 0.00232734 |
| 55 | RVE1 | 39879 | 29949 | 75.0997 | AAAAAAATATCTTA | 4 | 0.0003412 |
| 56 | At3g11280 | 2031 | 1353 | 66.6174 | ATCCTTATCTTCAT | 9 | 0.00360164 |
| 57 | AT5G61620 | 19901 | 9242 | 46.4399 | AATTTGGATAAGATT | 4 | 0.00302999 |
| 58 | AT3G10580 | 5180 | 1542 | 29.7683 | AATTAGGATAAGGTT | 2 | 0.00112477 |
| 59 | AT5G56840 | 31403 | 11591 | 36.9105 | AACCTTATCCATAAA | 6 | 0.00143684 |
| 60 | At5g05790 | 3605 | 1537 | 42.6352 | AACCTTATCTTCATC | 3 | 0.00155138 |
| 61 | CAMTA1 | 2776 | 1717 | 61.8516 | AAAGCGCGTGAA | 1 | 9.11E-05 |
| 62 | HAT2 | 14941 | 13427 | 89.8668 | ATTAATCATTA | 4 | 4.91E-04 |
| 63 | AGL63 | 15745 | 7549 | 47.9454 | TTCCAAAAATGGAAA | 6 | 4.17E-03 |
| 64 | ARF2 | 11654 | 6051 | 51.9221 | ATCCGACAAA | 9 | 1.82E-04 |
| 65 | TINY | 1775 | 639 | 36 | CTCCACCGACAAAAT | 1 | 0.00465732 |
| 66 | ERF5 | 846 | 489 | 57.8014 | CCACCGCCGCCGCCA | 1 | 0.000804042 |
| 67 | ERF15 | 25902 | 12754 | 49.2394 | CCTTAGCCGCCGCC | 4 | 2.76E-03 |
| 68 | AIL7 | 3535 | 2669 | 75.5021 | CGATTCCCGAG | 1 | 9.39E-04 |
| 69 | ERF10 | 4811 | 823 | 17.1066 | TCCACCGCCTCCGCCGCCGCC | 1 | 7.31E-05 |
| 70 | ABR1 | 11691 | 1743 | 14.9089 | GGCGGAAATGGCGGCGGCG | 1 | 0.00133472 |
| 71 | LEP | 546 | 167 | 30.5861 | TCCGCCGCCGCCACCGCCGCC | 1 | 0.000250877 |
| 72 | ERF2 | 726 | 213 | 29.3388 | CCGCCGCCTCCGCCGCCGCCA | 1 | 0.000143283 |
| 73 | TCP24 | 3352 | 1999 | 59.636 | GTTGGGACCACA | 1 | 0.0032299 |
| 74 | TCP3 | 1416 | 648 | 45.7627 | GTTGGGACCACAT | 1 | 7.78E-05 |
| 75 | TCP7 | 1002 | 877 | 87.525 | GTGGGACCCAC | 1 | 2.50E-05 |
| 76 | MYB27 | 8363 | 4259 | 50.9267 | AAAAAATTAGGTAAA | 2 | 0.00155854 |
| 77 | MYB57 | 12402 | 7833 | 63.1592 | TTTACCTAACTTTT | 6 | 1.45E-04 |
| 78 | MYB62 | 12953 | 4460 | 34.4322 | AAAAAAGTTAGGTAG | 3 | 0.000995587 |
| 79 | WRKY20 | 2213 | 1063 | 48.0343 | AACGTTGACTATT | 1 | 0.000369049 |
| 80 | WRKY29 | 18818 | 16438 | 87.3525 | AAAAGTCAACG | 1 | 0.00169441 |
| 81 | WRKY6 | 5323 | 534 | 10.0319 | CGTTGACTTTTTTTAACAT | 4 | 2.66E-07 |
| 82 | WRKY33 | 5593 | 4840 | 86.5367 | AAAAGTCAACG | 6 | 0.00203992 |
| 83 | WRKY65 | 20576 | 17810 | 86.5572 | AAAAGTCAACG | 1 | 0.000482904 |
| 84 | WRKY22 | 37563 | 17162 | 45.6886 | AAAAGTCAACGAT | 4 | 0.000326708 |
| 85 | WRKY55 | 9034 | 5724 | 63.3606 | AGCGTTGACTTT | 8 | 0.000786727 |
| 86 | WRKY31 | 3489 | 389 | 11.1493 | AATTGGTCAACGCTGGTT | 1 | 0.00208445 |
| 87 | WRKY70 | 8244 | 3749 | 45.4755 | AGCGTTGACTTTT | 8 | 0.000974108 |
| 88 | WRKY3 | 1287 | 1112 | 86.4025 | AAAAGTCAACG | 1 | 0.000449426 |
| 89 | WRKY28 | 13501 | 10654 | 78.9127 | AAGGTCAACGA | 5 | 0.00146025 |
| 90 | WRKY7 | 1926 | 1220 | 63.3437 | AGCGTTGACTTTTT | 1 | 0.00183075 |
| 91 | WRKY14 | 13405 | 6825 | 50.9138 | AAAAGTCAACGAT | 1 | 0.00185991 |
| 92 | WRKY24 | 18866 | 11792 | 62.504 | TCGTTGACTTTTTT | 4 | 0.000153134 |
| 93 | WRKY71 | 9533 | 8152 | 85.5135 | AAAAGTCAACG | 4 | 0.000285292 |
| 94 | WRKY50 | 13630 | 6424 | 47.1313 | CTTTGACTTTTTT | 3 | 7.94E-05 |
| 95 | WRKY27 | 23333 | 11001 | 47.1478 | ATCGTTGACTTTT | 5 | 1.92E-03 |
| 96 | SPL15 | 19981 | 10130 | 50.6982 | AAATTGTACGGACA | 1 | 0.00391891 |
| 97 | SPL9 | 41646 | 34721 | 83.3718 | TATCCGTACAA | 1 | 5.26E-05 |
| 98 | GATA19 | 202 | 192 | 95.0495 | CAGATCGGATT | 1 | 0.00124343 |
| 99 | GATA20 | 17519 | 17440 | 99.5491 | ATCGGATC | 6 | 0.00203506 |
| 100 | TGA4 | 10140 | 9561 | 94.2899 | TGACGTCATCA | 1 | 6.83E-05 |
| 101 | TGA3 | 3451 | 2669 | 77.3399 | GATGACGTCATCAT | 1 | 0.000326619 |
| 102 | bZIP44 | 2679 | 1031 | 38.4845 | GATGCTGACGTGGCA | 7 | 1.76E-03 |
| 103 | bZIP3 | 8295 | 2807 | 33.8397 | GATGCTGACGTGGCA | 6 | 0.000626708 |
| 104 | bZIP53 | 17446 | 5216 | 29.898 | GATGCTGACGTGGCA | 8 | 0.000150825 |
| 105 | bZIP52 | 16537 | 8034 | 48.582 | TTGACAGCTGGTCA | 3 | 0.00060522 |
| 106 | bZIP28 | 1726 | 786 | 45.5388 | AATGATGACGTGGCA | 3 | 1.72E-05 |
| 107 | bZIP48 | 5105 | 2717 | 53.2223 | GCCACGTCAGCATC | 9 | 0.000447777 |
| 108 | TGA10 | 5500 | 2667 | 48.4909 | GATGATGACGTCACT | 1 | 0.000199503 |
| 109 | TGA9 | 18677 | 17527 | 93.8427 | GATGACGTCAT | 1 | 0.00327323 |
| 110 | bZIP16 | 8404 | 3572 | 42.5036 | AATGGTCACGTGGCA | 8 | 0.00202097 |
| 111 | GBF3 | 16034 | 13481 | 84.0776 | CACGTGGCATA | 1 | 0.00123616 |
| 112 | TRP1 | 1042 | 80 | 7.67754 | ACCCTAAACCCTAAACCCTAA | 2 | 0.000273712 |
| 113 | AT1G72740 | 21483 | 5709 | 26.5745 | GGATTTAGGGTTTAG | 6 | 0.000361651 |
| 114 | TRP2 | 3036 | 272 | 8.95916 | AACCCTAAACCCTAAACCCTA | 2 | 1.80E-03 |
| 115 | bHLH80 | 5667 | 5136 | 90.63 | TGCAAGTTGCA | 8 | 0.00010231 |
| 116 | AT5G47660 | 12711 | 10854 | 85.3906 | ATGGTAAAAAA | 4 | 2.38E-05 |
| 117 | HDG1 | 11647 | 10291 | 88.3575 | GTAATTAATGC | 7 | 0.000558874 |
| 118 | IDD5 | 9121 | 2022 | 22.1686 | TTTTTGTCGTTTTGTG | 3 | 0.00060862 |
| 119 | IDD4 | 12546 | 3660 | 29.1726 | CACAAAACGACAAAAAA | 7 | 9.43E-05 |
| 120 | IDD7 | 4720 | 1036 | 21.9492 | CACAAAACGACAAAAAAA | 3 | 0.00307268 |
| 121 | ANL2 | 8312 | 7033 | 84.6126 | GCATTAATTAC | 6 | 4.57E-04 |
| 122 | DEAR3 | 12720 | 2884 | 22.673 | AATTGTCGGTGGTGGCA | 5 | 0.00280044 |
| 123 | SOL1 | 21836 | 10442 | 47.8201 | TTTTAAAATTTAAAT | 8 | 0.00158259 |
| 124 | FAR1 | 957 | 425 | 44.4096 | ACCTCACGCGCCTCC | 1 | 4.17E-06 |
| 125 | AT2G40260 | 21916 | 11147 | 50.8624 | TTAAAATATTCTTTT | 2 | 0.00434188 |
| 126 | MYB33 | 22513 | 7234 | 32.1325 | TAATAACCGTAAA | 8 | 3.92E-03 |
| 127 | MYB98 | 5558 | 4879 | 87.7834 | ATACGTTACAT | 1 | 1.54E-03 |
| 128 | MYB70 | 11564 | 10477 | 90.6001 | ATAACCGTTAT | 1 | 4.75E-03 |
| 129 | MYB73 | 9380 | 5699 | 60.7569 | TTAACGGTCAAA | 1 | 6.25E-05 |
| 130 | At1g74840 | 10169 | 8518 | 83.7644 | AATGGATAAGG | 5 | 2.99E-06 |
| 131 | At1g19000 | 29932 | 19505 | 65.1644 | AAATGGATAAGGTT | 9 | 0.000443634 |
| 132 | BPC1 | 17699 | 795 | 4.49178 | GAGAGAGAGAGAGAGAGAGAGAGA | 9 | 0.00231573 |
| 133 | E2FA | 7135 | 4864 | 68.171 | TGGCGCCAAA | 1 | 0.0002382 |
| 134 | GTL1 | 31474 | 21567 | 68.5232 | ATGGTTAAAAAT | 4 | 0.0023513 |
| 135 | AT3G10030 | 3648 | 1514 | 41.5022 | AGTTAACAGAAAT | 4 | 1.72E-03 |
| 136 | HSFA6B | 34926 | 18921 | 54.1745 | GCTTCTGGAAGC | 1 | 1.37E-08 |
| 137 | HSFC1 | 8354 | 4339 | 51.9392 | GCTTCCAGAAGC | 8 | 0.00293026 |
| 138 | LBD18 | 20264 | 11460 | 56.5535 | TCGCCGGAAAAT | 1 | 0.00051374 |
| 139 | NTL8 | 5913 | 2398 | 40.5547 | ATGAAGAAACCTA | 9 | 0.00264297 |
| 140 | AT1G19040 | 994 | 523 | 52.6157 | CTTGCGCCCCAAGTTA | 1 | 1.11E-03 |
| 141 | At5g04390 | 9238 | 8030 | 86.9236 | CAACTTCACTA | 3 | 0.00450835 |
| 142 | bHLH122 | 10050 | 6308 | 62.7662 | TTAGCAACTTGCAT | 4 | 0.00231664 |
| 143 | bZIP18 | 14191 | 12230 | 86.1814 | TGACAGCTGGT | 4 | 0.00140816 |
| 144 | GATA4 | 1611 | 1306 | 81.0677 | ATAGATCTAT | 1 | 0.00134676 |
| 145 | GRF9 | 2374 | 1007 | 42.4179 | TTGTCAGAAAATACA | 4 | 2.36E-04 |
| 146 | HSFB3 | 1171 | 825 | 70.4526 | AAGCTTCTAGAAG | 1 | 6.26E-05 |
| 147 | MYB116 | 8961 | 3387 | 37.7971 | TCTGCCTAACTTT | 8 | 3.82E-03 |
| 148 | MYB121 | 2067 | 1356 | 65.6023 | TTATACCTAACTTT | 1 | 0.00388283 |
| 149 | MYB61 | 17756 | 5004 | 28.182 | CTCCACCTACCAT | 7 | 0.000105934 |
| 150 | MYB67 | 19668 | 674 | 3.42689 | TCCACCTAACATTACCACTAC | 4 | 0.00109708 |
| 151 | MYB74 | 3061 | 1764 | 57.6282 | TTTCACCTACCTTT | 1 | 6.46E-05 |
| 152 | MYB83 | 14502 | 10471 | 72.2038 | CCACCAACCAC | 3 | 0.00116736 |
| 153 | NLP7 | 867 | 294 | 33.91 | ATTTGGCTTTTCGGA | 3 | 5.65E-05 |
| 154 | VIP1 | 1523 | 938 | 61.589 | TTGACAGCTGTACA | 1 | 2.82E-04 |
| 155 | MYB107 | 13462 | 5061 | 37.5947 | TTCACCAAACATA | 9 | 0.00225118 |
| 156 | MYB99 | 4060 | 2611 | 64.3103 | TTCACCTAAC | 6 | 0.00144785 |
| 157 | SMB | 40054 | 32476 | 81.0805 | TACAAGCAATC | 1 | 1.62E-05 |
| 158 | ERF115 | 12101 | 4646 | 38.3935 | CGTCGCCGCCGCAGC | 1 | 0.0028817 |
| 159 | BAM8 | 4589 | 3764 | 82.0222 | TTCACACGTGCGAA | 1 | 0.00150621 |
| 160 | LBD13 | 3715 | 1023 | 27.537 | CCTCCACCGTCTC | 9 | 0.00499397 |
| 161 | LOB | 1461 | 679 | 46.475 | CCGCCGCCGCCGCCGCC | 1 | 0.000376676 |
| 162 | MYB63 | 2666 | 972 | 36.4591 | TCCACCTACCACC | 4 | 0.00224502 |
| 163 | MYB93 | 7813 | 4142 | 53.0142 | TCTACCAAACCAAA | 9 | 0.00047858 |
| 164 | MYB44 | 15418 | 9299 | 60.3126 | TTAACGGTCAAA | 1 | 0.00427091 |
| 165 | MYB49 | 9961 | 3937 | 39.5241 | TTTTACCTAACTATT | 3 | 0.0025348 |
| 166 | MYB88 | 15118 | 4590 | 30.3612 | ACACGCTCCTCCA | 2 | 0.000150039 |
| 167 | MYB96 | 1147 | 403 | 35.1351 | AACTCCCAACTACCAAC | 1 | 0.00271496 |
| 168 | AT5G05550 | 18951 | 11005 | 58.0708 | ATCTCCGGCGATG | 7 | 2.89E-06 |
| 169 | MYB13 | 2733 | 596 | 21.8075 | ATACCCACCAACCAAA | 6 | 0.00360942 |
| 170 | MYB58 | 21702 | 5402 | 24.8917 | ATCTCCACCAACCCCAA | 5 | 8.48E-08 |
| 171 | PIF1 | 5491 | 3871 | 70.4971772 | AGAAGGTCACGTGG | 1 | 0.00420378 |
| 172 | SOC1 | 2273 | 1830 | 80.51033876 | TTTTTCCATTTTTGG | 1 | 0.00254781 |
| 173 | SVP | 4330 | 226 | 5.219399538 | ATTACCAAAAAAGGAAAGAA | 3 | 0.00364276 |
| 174 | FHY3 | 444 | 294 | 66.21621622 | CTCACGCGCTCA | 9 | 0.0030724 |
| 175 | PIF4 | 33969 | 30572 | 89.99970561 | CACGTGGC | 1 | 2.98E-06 |
| 176 | PIF5 | 3378 | 3040 | 89.99407934 | TCACGTGG | 1 | 2.55E-03 |
| 177 | SEP3 | 24925 | 24566 | 98.55967904 | TCCATTTTTGG | 8 | 0.000543462 |
| 178 | ABF1 | 22241 | 16213 | 72.89690212 | ACACGTGGCATA | 1 | 3.72E-03 |
| 179 | LFY | 10818 | 366 | 3.383250139 | TATTGACCGCCGGTCAATA | 3 | 1.42E-04 |
| 180 | ABF3 | 37937 | 28718 | 75.69918549 | CACGTGTCAT | 1 | 1.49E-04 |
| 181 | ABI5 | 2798 | 2123 | 75.87562545 | TGACACGTGG | 1 | 0.00127782 |
| 182 | AP1 | 5108 | 4180 | 81.83241973 | CCAAAAAAGGAAA | 3 | 0.00176705 |
| 183 | CCA1 | 1206 | 1205 | 99.91708126 | AAATATCT | 4 | 0.000273669 |
| 184 | KAN1 | 20007 | 16005 | 79.99700105 | AGATATTC | 1 | 2.41E-03 |
| 185 | MYB3 | 20813 | 19780 | 95.03675587 | AGGTAGGTAGA | 6 | 0.000488737 |
| 186 | HAT22 | 31291 | 27868 | 89.06075229 | ATCAATAATTGAA | 4 | 2.71E-06 |
| 187 | DREB2A | 3797 | 154 | 4.055833553 | TGGTTATGTCGGTGGAGGTGG | 2 | 4.50E-05 |
| 188 | WRKY33 | 7355 | 7266 | 98.78993882 | AAAAGTCAACG | 9 | 8.87E-05 |
| 189 | BZIP28 | 1102 | 651 | 59.07441016 | AATGATGACGTGGCA | 7 | 1.83E-06 |
| 190 | GBF3 | 39592 | 30881 | 77.99808042 | CACGTGGCATA | 1 | 0.00302175 |
| 191 | ABF4 | 31851 | 22773 | 71.49854008 | TGCCACGTCACC | 1 | 0.00278147 |
| 192 | GBF2 | 31090 | 22695 | 72.99774847 | GTGACGTGGCAAA | 1 | 2.83E-05 |
| 193 | HSFA6A | 8898 | 5463 | 61.39581929 | GAAGCTTCTAGAA | 9 | 0.0039441 |
| 194 | ERF115 | 11318 | 2293 | 20.25976321 | CGTCGCCGCCGCAGC | 1 | 0.00363688 |
| 195 | MYB44 | 34581 | 29991 | 86.7268153 | TTAACGGTCAAA | 1 | 0.00427091 |
| 196 | AT5G04760 | 30418 | 30197 | 99.27345651 | ATCTTATCCTT | 3 | 0.000253518 |
| 197 | ZAT6 | 20754 | 20124 | 96.96444059 | TTAATGATTGAT | 8 | 4.35E-05 |
Sheet 7: Information of experimentally derived motif from Plant Cistrome database of DAP-seq origin
| S.No. | TF | Total peaks | Covered peaks | Coverage (%) | Consensus | p-value |
|---|---|---|---|---|---|---|
| 1 | AT5G60130 | 19106 | 1701 | 8.90296242 | AAGCAAAAAATAAGCAAAA | 0.00491599 |
| 2 | FUS3 | 3266 | 1868 | 57.19534599 | ATATACATGCATGCA | 4.53E-06 |
| 3 | NGA4 | 2235 | 425 | 19.01565996 | CCTGTTCAGGTGATTT | 0.00166732 |
| 4 | VRN1 | 7028 | 4 | 0.056915196 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTC | 0.000730868 |
| 5 | ABR1 | 11691 | 1743 | 14.90890429 | GGCGGAAATGGCGGCGGCG | 2.92E-05 |
| 6 | AIL7 | 3535 | 2669 | 75.50212164 | CGATTCCCGAG | 0.000879391 |
| 7 | AT1G01250 | 725 | 303 | 41.79310345 | GATATTGTCGGTGGT | 0.00407569 |
| 8 | AT1G12630 | 18211 | 1292 | 7.094613146 | CGCCGCCGCCTTCGCCGACAA | 2.52E-05 |
| 9 | At1g19210 | 22451 | 9646 | 42.96467863 | ATGGACGGTGGAGG | 0.00164144 |
| 10 | At1g22810 | 8437 | 3045 | 36.09102762 | GGCGGTGGCGGTGGA | 0.000252257 |
| 11 | AT1G28160 | 10300 | 4618 | 44.83495146 | TGGCGGCGGCGGTGG | 0.00383144 |
| 12 | At1g36060 | 4035 | 2054 | 50.90458488 | ATGGTCGGTGGAGG | 2.72E-05 |
| 13 | AT1G44830 | 1635 | 669 | 40.91743119 | CCACCTCCACCGCCA | 0.00465732 |
| 14 | AT1G71450 | 14277 | 1206 | 8.447152763 | CACCGCCGCCGCCACCGCCA | 0.00211046 |
| 15 | AT1G77200 | 27117 | 12562 | 46.32518346 | CCACCGACAAATTC | 0.00445287 |
| 16 | At2g33710 | 18654 | 6794 | 36.42114292 | CCTCCGCCGCCGCCA | 3.17E-05 |
| 17 | At2g44940 | 5163 | 1591 | 30.81541739 | AATTGTCGGTGGAGT | 0.000151555 |
| 18 | AT3G16280 | 13312 | 5926 | 44.51622596 | TATTGTCGGTGGAG | 0.00401278 |
| 19 | AT3G60490 | 9000 | 4283 | 47.58888889 | AATATTGTCGGTGG | 0.00238675 |
| 20 | At4g16750 | 15677 | 4073 | 25.98073611 | CACCGCCACCGACAC | 0.000104656 |
| 21 | AT4G18450 | 561 | 321 | 57.21925134 | GCGGCGGCGGCGGCGGC | 0.000117875 |
| 22 | At4g31060 | 7514 | 497 | 6.614319936 | CCACCGACAAAACCACCACC | 0.00288674 |
| 23 | At4g32800 | 3779 | 712 | 18.84096322 | CTCCACCGACAATTTC | 0.000612668 |
| 24 | At5g18450 | 2405 | 1036 | 43.07692308 | GGTGGAGGCGGTGGA | 7.25E-05 |
| 25 | At5g65130 | 1992 | 961 | 48.24297189 | CCACCGCCGCCGCCA | 0.00110258 |
| 26 | CBF1 | 18542 | 10242 | 55.23675979 | TTCGCCGACATC | 0.00391828 |
| 27 | CBF2 | 14034 | 3875 | 27.61151489 | TCTTTGCCGACATCA | 0.00172184 |
| 28 | CBF3 | 12163 | 6173 | 50.75228151 | CTTCACCGACATAA | 0.00184183 |
| 29 | CBF4 | 29710 | 8401 | 28.27667452 | TGATGTCGGCGAA | 0.00193623 |
| 30 | CEJ1 | 30321 | 7903 | 26.06444378 | CCACCTCCACCGACA | 0.00151031 |
| 31 | CRF10 | 6410 | 3162 | 49.32917317 | CCGCCGCCGCCGCCA | 0.00184638 |
| 32 | CRF4 | 2482 | 774 | 31.18452861 | CCTCCGCCGCCGCCGCCGCCG | 0.00495489 |
| 33 | DDF1 | 2395 | 910 | 37.99582463 | TGATGTCGGCGACGA | 0.00326973 |
| 34 | DDF2 | 326 | 142 | 43.55828221 | TGATGTCGGCGAAGA | 0.000975699 |
| 35 | DEAR2 | 23238 | 6913 | 29.74868749 | CCACCGCCACCGACA | 2.07E-06 |
| 36 | DEAR3 | 12720 | 2884 | 22.67295597 | AATTGTCGGTGGTGGCA | 0.000561833 |
| 37 | DEAR5 | 2274 | 227 | 9.98240985 | CTTCATCACCACCGACAAT | 0.00141631 |
| 38 | DREB19 | 15090 | 3481 | 23.06825712 | CACCTCCACCGACATTA | 2.21E-06 |
| 39 | ERF105 | 14787 | 5891 | 39.83904781 | GTGGCGGCGGCGGAG | 5.98E-05 |
| 40 | ERF10 | 4811 | 823 | 17.10663064 | TCCACCGCCTCCGCCGCCGCC | 0.000486218 |
| 41 | ERF115 | 12101 | 4680 | 38.67448971 | CCGCCGCCACCGCCG | 0.000722211 |
| 42 | ERF11 | 7672 | 3149 | 41.04535975 | CCACCGCCGCCGCCA | 0.00373966 |
| 43 | ERF13 | 9556 | 2914 | 30.49393051 | CTCCGCCGCCATTTC | 0.00115298 |
| 44 | ERF15 | 25902 | 7857 | 30.33356498 | CGCCTCCGCCGCCGC | 0.00109119 |
| 45 | ERF2 | 726 | 213 | 29.33884298 | CCGCCGCCTCCGCCGCCGCCA | 4.48E-05 |
| 46 | ERF38 | 5518 | 1644 | 29.79340341 | TGTCGGTGGAGGTGG | 0.000502342 |
| 47 | ERF3 | 3321 | 1639 | 49.35260464 | TGGCGGCGGCGGTGG | 0.00190022 |
| 48 | ERF4 | 15193 | 5778 | 38.03067202 | CCTCCGCCGCCATTT | 0.00103313 |
| 49 | ERF5 | 846 | 489 | 57.80141844 | CCACCGCCGCCGCCA | 0.000105279 |
| 50 | ERF7 | 17694 | 1966 | 11.11111111 | CCACCGCCTCCGCCGCCGCCA | 0.000436962 |
| 51 | ERF8 | 12829 | 4692 | 36.57338842 | TCCTCCGCCGCCGCC | 0.000552481 |
| 52 | ESE1 | 1512 | 947 | 62.63227513 | CGGCGGCGGCGGCGG | 3.15E-05 |
| 53 | ESE3 | 9328 | 1924 | 20.62607204 | CGGAGGTGGCGGCGGAGGA | 9.72E-05 |
| 54 | LEP | 546 | 167 | 30.58608059 | TCCGCCGCCGCCACCGCCGCC | 0.000585707 |
| 55 | PLT3 | 541 | 187 | 34.56561922 | CCTCGGGATTCGTGC | 0.000514223 |
| 56 | PUCHI | 2691 | 1280 | 47.56596061 | CCGCCGCCGCCGCCA | 0.00348221 |
| 57 | Rap210 | 12600 | 3707 | 29.42063492 | TTTTGTCGGCAAAGG | 0.00189035 |
| 58 | RAP211 | 16230 | 4424 | 27.25816389 | CCGCCGCCGCCGGCG | 0.00374906 |
| 59 | RAP212 | 6571 | 969 | 14.74661391 | CGGCGGAAATGGCGGCGGAGG | 4.08E-06 |
| 60 | RAP21 | 2103 | 848 | 40.3233476 | GTGTCGGTGGTGG | 0.000165964 |
| 61 | RAP26 | 8572 | 3450 | 40.24731685 | TGGCGGCGGCGGAGG | 0.00487556 |
| 62 | RRTF1 | 8037 | 3398 | 42.27945751 | GAAATTGCGGCGGCG | 6.65E-06 |
| 63 | TINY | 1775 | 639 | 36 | CTCCACCGACAAAAT | 0.00124645 |
| 64 | MP | 26659 | 14020 | 52.59011966 | GCCGACAAAA | 6.60E-05 |
| 65 | ARF2 | 11654 | 6051 | 51.92208684 | ATCCGACAAA | 0.000730672 |
| 66 | AT1G20910 | 10683 | 9198 | 86.09941028 | AAATTTAATTT | 0.00150977 |
| 67 | BPC1 | 17699 | 795 | 4.491779197 | GAGAGAGAGAGAGAGAGAGAGAGA | 0.00285794 |
| 68 | BAM8 | 4589 | 3043 | 66.31074308 | TCACACGTGTGAACT | 0.0010857 |
| 69 | bHLH10 | 9144 | 4087 | 44.6959755 | GAATGTGTCGGTGG | 0.000615864 |
| 70 | bHLH122 | 10050 | 6308 | 62.76616915 | TTAGCAACTTGCAT | 0.000102413 |
| 71 | bHLH28 | 8905 | 5351 | 60.08983717 | AATTGTACGGAC | 7.00E-06 |
| 72 | bHLH31 | 1829 | 106 | 5.795516676 | GAGGTGGCACGTGGCATTCAC | 0.000858712 |
| 73 | bHLH80 | 5667 | 5136 | 90.62996294 | TGCAAGTTGCA | 0.000117768 |
| 74 | BIM1 | 647 | 486 | 75.11591963 | CACGTGACATCCAC | 0.000359122 |
| 75 | BIM2 | 40836 | 11430 | 27.99000882 | CACGTGACTTTCACG | 0.00425213 |
| 76 | ABF2 | 2299 | 605 | 26.31578947 | AAAAATGCCACGTGACCA | 0.000140965 |
| 77 | ABI5 | 9233 | 1705 | 18.46637063 | AAATGGTGACGTGGCAGT | 0.00212496 |
| 78 | AREB3 | 4617 | 2231 | 48.32142084 | AATGGACACGTGGCA | 0.00091545 |
| 79 | bZIP16 | 8404 | 3572 | 42.50356973 | AATGGTCACGTGGCA | 0.00271717 |
| 80 | bZIP18 | 14191 | 12282 | 86.54781199 | ACCAGCTGTCA | 0.00372424 |
| 81 | bZIP28 | 1726 | 786 | 45.53881808 | AATGATGACGTGGCA | 0.00375453 |
| 82 | bZIP3 | 8295 | 2807 | 33.83966245 | GATGCTGACGTGGCA | 0.00195421 |
| 83 | bZIP44 | 2679 | 1031 | 38.48450915 | GATGCTGACGTGGCA | 0.00264297 |
| 84 | bZIP48 | 5105 | 2717 | 53.22233105 | GCCACGTCAGCATC | 0.000372035 |
| 85 | bZIP50 | 22131 | 6867 | 31.02887353 | TGATGACGTCATCTTTT | 0.00160215 |
| 86 | bZIP52 | 16537 | 8034 | 48.58196771 | TTGACAGCTGGTCA | 0.00484464 |
| 87 | bZIP53 | 17446 | 5216 | 29.89797088 | GATGCTGACGTGGCA | 0.000169538 |
| 88 | bZIP68 | 9044 | 2877 | 31.81114551 | TGCCACGTCAGCATC | 0.00386836 |
| 89 | GBF3 | 16034 | 5027 | 31.35212673 | TGCCACGTCAGCATT | 0.00124458 |
| 90 | GBF5 | 6575 | 2498 | 37.99239544 | AATGCTGACGTGGCA | 0.00102133 |
| 91 | GBF6 | 9894 | 2791 | 28.20901556 | TGCCACGTCAGCATC | 0.00019017 |
| 92 | TGA10 | 5500 | 2667 | 48.49090909 | GATGATGACGTCACT | 0.000798543 |
| 93 | TGA1 | 12377 | 6567 | 53.05809162 | AATGATGACGTCATC | 0.000483463 |
| 94 | TGA2 | 16587 | 694 | 4.183999518 | AATGACGTCATCATTATTGT | 7.93E-06 |
| 95 | TGA3 | 3451 | 2669 | 77.33990148 | GATGACGTCATCAT | 0.00138835 |
| 96 | TGA4 | 10140 | 9561 | 94.28994083 | TGACGTCATCA | 0.000458564 |
| 97 | TGA5 | 2604 | 2030 | 77.95698925 | AATGACGTCATCAT | 0.00192131 |
| 98 | TGA6 | 28254 | 13111 | 46.40404898 | AATGACGTCATCATT | 0.00148448 |
| 99 | TGA9 | 18677 | 17527 | 93.84269422 | GATGACGTCAT | 0.00180227 |
| 100 | VIP1 | 1523 | 1121 | 73.60472751 | TTGACAGCTGTA | 0.000893241 |
| 101 | At1g78700 | 5147 | 4692 | 91.15989897 | CGCACGTGTGG | 5.46E-05 |
| 102 | At4g18890 | 865 | 844 | 97.57225434 | CGCACGTGTGA | 0.000299943 |
| 103 | At4g36780 | 6039 | 5583 | 92.44908097 | TCACACGTGCG | 0.000197593 |
| 104 | BZR1 | 1018 | 80 | 7.858546169 | CCACACGTGCGACTTTTTTT | 0.000675588 |
| 105 | Adof1 | 11744 | 1599 | 13.61546322 | AAAAAGAAAAAGTAAAAAAAA | 0.000394283 |
| 106 | AT1G47655 | 7940 | 7040 | 88.66498741 | AGAAAAAGTAA | 0.00207979 |
| 107 | At1g64620 | 34001 | 4040 | 11.88200347 | AAAAAAAAAAAAGTAAAAA | 0.00412504 |
| 108 | AT1G69570 | 13762 | 390 | 2.833890423 | TTTTCACTTTTTCTTTTTTTTTTTTTT | 0.0047197 |
| 109 | AT2G28810 | 49623 | 5512 | 11.10775245 | TTTTTTTTTTTTTACTTTTTT | 1.14E-06 |
| 110 | At3g45610 | 34597 | 4522 | 13.07049744 | TTTACTTTTTGCTTTTTTT | 0.000826851 |
| 111 | AT3G52440 | 30433 | 26628 | 87.49712483 | CAAAAAGTTAA | 0.000233957 |
| 112 | At4g38000 | 11840 | 114 | 0.962837838 | TTTTTTTTTTTTTTTTTTTTACTTTTTT | 0.00172251 |
| 113 | AT5G02460 | 45371 | 4866 | 10.72491239 | AAAAAAGTAAAAAAAAAAAAA | 0.000314108 |
| 114 | At5g62940 | 29881 | 3142 | 10.515043 | AAAAAAAAGAAAAAGTAAAAA | 6.54E-05 |
| 115 | AT5G66940 | 40237 | 146 | 0.362850113 | TTTTTTTTTTTTTTTACTTTTTTTTTTTT | 0.00237475 |
| 116 | CDF3 | 22820 | 2993 | 13.11568799 | AAAAAAGAAAAAGTGAAAA | 2.09E-07 |
| 117 | COG1 | 11354 | 1424 | 12.54183548 | AAAAAAAAGAAAAAGTGAAAA | 0.000504084 |
| 118 | DAG2 | 24338 | 21400 | 87.92834251 | CAAAAAGTAAA | 0.00420165 |
| 119 | dof24 | 27058 | 11145 | 41.18929707 | TTTTTTTACTTTTTG | 0.00266762 |
| 120 | dof42 | 801 | 133 | 16.60424469 | AAAAAAAAAAGGCAAAAAA | 0.000819903 |
| 121 | dof43 | 19070 | 16974 | 89.00891453 | AAAGCTACTTT | 0.00357571 |
| 122 | dof45 | 24304 | 14673 | 60.37277814 | GAAAAAGTAACTTT | 0.000455133 |
| 123 | OBP1 | 23458 | 2617 | 11.15610879 | TTTTTTTTTTTTTTACTTTTT | 0.00356383 |
| 124 | OBP3 | 21664 | 1963 | 9.061115214 | TTTACTTTTTTTTTTTTTTTT | 0.00439065 |
| 125 | OBP4 | 27993 | 18204 | 65.03054335 | CAAAAAGTTAAAAA | 0.00202278 |
| 126 | GATA11 | 3149 | 1967 | 62.46427437 | AGATCTAGATCTGAA | 4.75E-05 |
| 127 | GATA19 | 202 | 192 | 95.04950495 | CAGATCGGATT | 0.00282092 |
| 128 | GATA1 | 970 | 316 | 32.57731959 | ATGGTGATGATGATG | 0.000144948 |
| 129 | GATA20 | 17519 | 17440 | 99.54906102 | ATCGGATC | 0.00357505 |
| 130 | GATA4 | 1611 | 1052 | 65.30105525 | TTTCAGATCTGAATC | 0.000940301 |
| 131 | CRC | 9178 | 9152 | 99.71671388 | TATGATTA | 0.00203506 |
| 132 | AT2G15740 | 3257 | 2971 | 91.21891311 | AACGATGACGA | 0.000349564 |
| 133 | At3g60580 | 11065 | 10987 | 99.29507456 | TGTAGTAG | 4.47E-05 |
| 134 | AT4G26030 | 11315 | 974 | 8.608042422 | CAATAATTTCACCAACCAT | 0.000712653 |
| 135 | At5g04390 | 9238 | 8001 | 86.60965577 | TAGTGAAGTTG | 2.78E-05 |
| 136 | AT5G22990 | 4898 | 2063 | 42.11923234 | TCGTCATCGTTTTCG | 0.00210867 |
| 137 | At5g66730 | 7624 | 2769 | 36.31951731 | AAGAAAACGACAAAAAA | 0.0027608 |
| 138 | AtIDD11 | 3281 | 447 | 13.62389515 | TTTTTTTTTTTGTCTTTTTCT | 3.20E-05 |
| 139 | IDD4 | 12546 | 3660 | 29.17264467 | CACAAAACGACAAAAAA | 0.000556861 |
| 140 | IDD5 | 9121 | 2022 | 22.16862186 | TTTTTGTCGTTTTGTG | 0.000216926 |
| 141 | IDD7 | 4720 | 1036 | 21.94915254 | CACAAAACGACAAAAAAA | 0.000258777 |
| 142 | JKD | 2160 | 221 | 10.23148148 | TTTTTTTTTGTCGTTTTCTG | 0.00121363 |
| 143 | MGP | 3771 | 346 | 9.17528507 | CAGAAAACGACAAAAAAAAA | 0.000833907 |
| 144 | SGR5 | 18969 | 8459 | 44.59381095 | AAAAAAAGACAAAAA | 0.00148843 |
| 145 | STOP1 | 1356 | 194 | 14.30678466 | CCTTCCCCAGATAAAACCT | 0.000758909 |
| 146 | STZ | 36348 | 31478 | 86.60173875 | CACTTTCACTA | 1.45E-05 |
| 147 | WIP5 | 8396 | 2451 | 29.19247261 | TACCTGGAGAACATA | 0.00228192 |
| 148 | At5g08750 | 968 | 360 | 37.19008264 | ACCTCAGCCGCAAAA | 0.00182073 |
| 149 | AT5G63260 | 18676 | 16875 | 90.35660741 | GAAAAAGTTAA | 0.000524817 |
| 150 | CAMTA1 | 2776 | 1717 | 61.85158501 | AAAGCGCGTGAA | 0.00123224 |
| 151 | CAMTA5 | 773 | 163 | 21.08667529 | CCGTGTAAAAAACGCGTG | 0.000359748 |
| 152 | AT2G20110 | 30396 | 12448 | 40.95275694 | TTTTAAATTTTTTAA | 0.00132961 |
| 153 | SOL1 | 21836 | 10442 | 47.82011357 | TTTTAAAATTTAAAT | 2.99E-06 |
| 154 | TCX2 | 24443 | 11442 | 46.81094792 | ATTTAAAATTCAAAT | 0.00171694 |
| 155 | DEL2 | 1401 | 85 | 6.067094932 | TTATTTTTTTGGCGGGAAAA | 0.00269597 |
| 156 | E2FA | 7135 | 4014 | 56.25788367 | ATTTGGCGCCAAAA | 0.00100203 |
| 157 | EIN3 | 1209 | 535 | 44.25144748 | GTCTAGATTCAATGA | 1.51E-07 |
| 158 | FAR1 | 957 | 425 | 44.40961338 | ACCTCACGCGCCTCC | 0.00155138 |
| 159 | At1g25550 | 15049 | 14167 | 94.13914546 | GGAATCTTCTA | 0.000388821 |
| 160 | AT1G49560 | 20976 | 20252 | 96.54843631 | ATCAAAGATTC | 0.000504352 |
| 161 | At1g68670 | 11676 | 6697 | 57.35697157 | GAATCAAAGATTC | 0.00131784 |
| 162 | At2g01060 | 21429 | 11698 | 54.58957488 | AAAAGAATATTCC | 1.67E-06 |
| 163 | At2g03500 | 12731 | 12091 | 94.97290079 | AGAATATTCCA | 0.00107169 |
| 164 | AT2G20400 | 6113 | 17 | 0.278095861 | CGAATATTCCTTTATTTCATCTGTATAAT | 0.00104865 |
| 165 | AT2G40260 | 21916 | 11147 | 50.86238365 | TTAAAATATTCTTTT | 0.00298174 |
| 166 | At3g04030 | 22876 | 17261 | 75.45462493 | AAAAGAATCTTCTT | 0.0035638 |
| 167 | At3g12730 | 10750 | 6139 | 57.10697674 | AAAGGGAATATTCC | 0.00211723 |
| 168 | At3g24120 | 52038 | 51887 | 99.70982743 | AGAATCTG | 0.000425009 |
| 169 | AT4G37180 | 19680 | 18245 | 92.70833333 | AAAAAGATTCC | 0.00484464 |
| 170 | At5g29000 | 4704 | 2729 | 58.01445578 | AAAGAATATTCCAAT | 0.00220332 |
| 171 | AT5G45580 | 42093 | 31932 | 75.8605944 | AAAAGAATATTCTT | 0.000209387 |
| 172 | AT4G00250 | 11654 | 5593 | 47.99210571 | AACCTTATCCATT | 0.00111434 |
| 173 | AtGRF6 | 6529 | 4056 | 62.12283658 | GAAAATGTGTCAGA | 0.00140275 |
| 174 | GRF9 | 2374 | 1007 | 42.41786015 | TTGTCAGAAAATACA | 0.00316257 |
| 175 | ANL2 | 8312 | 7033 | 84.61260828 | GCATTAATTAC | 0.000235686 |
| 176 | ATHB15 | 6538 | 3675 | 56.20985011 | AAAAGTAATGATTAC | 0.00411674 |
| 177 | ATHB21 | 22139 | 13180 | 59.5329509 | ACACCAATAATTGG | 0.00089564 |
| 178 | ATHB40 | 51300 | 5936 | 11.5711501 | AAACCAATAATTGAAAATTAT | 0.000261185 |
| 179 | ATHB53 | 17471 | 16026 | 91.72915116 | CCAATAATTGA | 0.00291234 |
| 180 | ATHB5 | 46401 | 44187 | 95.22855111 | TCAATAATTGA | 0.000143019 |
| 181 | LMI1 | 41115 | 19570 | 47.59820017 | TTCAATTATTGGT | 0.000234857 |
| 182 | PHV | 10285 | 5093 | 49.51871658 | GTAATCATTACTTTT | 0.00252777 |
| 183 | HAT2 | 14941 | 13427 | 89.86680945 | ATTAATCATTA | 0.00378723 |
| 184 | ATHB13 | 23232 | 21543 | 92.72985537 | TCAATAATTAA | 0.00422566 |
| 185 | ATHB18 | 5366 | 5001 | 93.19791278 | TCAATCATTGG | 0.00238979 |
| 186 | ATHB20 | 5911 | 5562 | 94.09575368 | TCAATAATTGA | 4.57E-06 |
| 187 | ATHB6 | 9807 | 9400 | 95.84990313 | CAATCATTTAT | 0.00340579 |
| 188 | ATHB7 | 10616 | 6912 | 65.10926903 | TCAATGATTGATT | 0.000316432 |
| 189 | HDG1 | 11647 | 10291 | 88.35751696 | GTAATTAATGC | 0.00373864 |
| 190 | PDF2 | 2160 | 1285 | 59.49074074 | AAAAGAATATTCCAA | 0.000729868 |
| 191 | HSF21 | 756 | 429 | 56.74603175 | TCTAGAAGCTTCTAGAAGC | 0.00435798 |
| 192 | HSF3 | 21814 | 19316 | 88.54863849 | GAAGCTTCTAGA | 0.000716423 |
| 193 | HSF6 | 1227 | 1134 | 92.4205379 | GAAGCTTCTAGA | 0.00144832 |
| 194 | HSF7 | 4474 | 4047 | 90.45596781 | TTCTAGAAGCTTCT | 0.00474108 |
| 195 | HSFA6B | 34926 | 18064 | 51.72078108 | AGAAGCTTCTAGAAG | 0.000749524 |
| 196 | HSFB3 | 1171 | 807 | 68.91545687 | CTTCTAGAAGCTTCT | 3.11E-07 |
| 197 | HSFC1 | 8354 | 7171 | 85.83911898 | CTAGAAGCTTC | 0.000415443 |
| 198 | ASL18 | 6328 | 2820 | 44.56384324 | CCGGAAAATCCGGAG | 2.94E-05 |
| 199 | LBD13 | 3715 | 1602 | 43.12247645 | CGGCGGAAATTGCGGCG | 0.00497509 |
| 200 | LBD18 | 20264 | 1866 | 9.20844848 | TTTCCTCCGGATTTTCCGG | 0.00358657 |
| 201 | LBD19 | 23977 | 8522 | 35.5423948 | CCGGAAAATCCGGAG | 9.15E-07 |
| 202 | LBD23 | 1451 | 519 | 35.76843556 | AGCGTATTTTACGCT | 0.0010164 |
| 203 | LBD2 | 24314 | 9308 | 38.282471 | CCGAAAAAATCGGAG | 7.73E-05 |
| 204 | LOB | 1461 | 428 | 29.29500342 | TCCGCCGCCGCCTCCGCCGCC | 0.00491626 |
| 205 | AGL15 | 2257 | 453 | 20.07089056 | CTTTCCTTTTTTGGAAAGT | 1.00E-05 |
| 206 | AGL63 | 15745 | 7549 | 47.94537949 | TTCCAAAAATGGAAA | 0.00196215 |
| 207 | SVP | 20687 | 7597 | 36.72354619 | ACTTTCCTTTTTTGG | 1.71E-05 |
| 208 | AT5G23930 | 5551 | 903 | 16.26733922 | CGCCGCCGCCACCGCCGCCG | 0.00193039 |
| 209 | At1g19000 | 29932 | 19505 | 65.16437258 | AAATGGATAAGGTT | 0.00434188 |
| 210 | At1g49010 | 28552 | 9045 | 31.67904175 | ATTAACCTTATCCTC | 0.000162186 |
| 211 | AT1G72740 | 21483 | 5971 | 27.79406973 | CCTAAACCCTAAATC | 0.00164165 |
| 212 | At1g74840 | 10169 | 8518 | 83.76438195 | AATGGATAAGG | 1.24E-06 |
| 213 | At3g09600 | 29529 | 22369 | 75.75264994 | AGATATTTTTTT | 0.000274278 |
| 214 | AT3G10113 | 27249 | 13143 | 48.23296268 | TAAAAAAATATCTTA | 0.00232903 |
| 215 | AT3G10580 | 5180 | 1542 | 29.76833977 | AATTAGGATAAGGTT | 0.000427741 |
| 216 | At3g11280 | 2031 | 1353 | 66.61742984 | ATCCTTATCTTCAT | 0.000463201 |
| 217 | At4g01280 | 24503 | 22171 | 90.48279802 | AGATATTTTTT | 0.00165137 |
| 218 | At5g05790 | 3605 | 1537 | 42.63522885 | AACCTTATCTTCATC | 3.00E-05 |
| 219 | At5g08520 | 12645 | 5323 | 42.09569 | AATATGGATAAGG | 0.00397994 |
| 220 | At5g47390 | 17465 | 6980 | 39.96564558 | ATAACCTTATCCATA | 2.56E-06 |
| 221 | At5g52660 | 19533 | 18010 | 92.20293862 | AGATATTTTTT | 1.48E-07 |
| 222 | AT5G56840 | 31403 | 11591 | 36.91048626 | AACCTTATCCATAAA | 0.000432347 |
| 223 | At5g58900 | 18957 | 7222 | 38.09674527 | TATGGATAAGGTTAT | 0.00212914 |
| 224 | AT5G61620 | 19901 | 9242 | 46.43987739 | AATTTGGATAAGATT | 5.45E-05 |
| 225 | EPR1 | 52997 | 40203 | 75.85901089 | AAAAATATCTTA | 1.91E-05 |
| 226 | LCL1 | 8370 | 6621 | 79.10394265 | AGATATTTTT | 0.00014989 |
| 227 | LHY1 | 33488 | 28104 | 83.92259914 | AGATATTTTT | 0.00213267 |
| 228 | RVE1 | 39879 | 29949 | 75.09967652 | AAAAAAATATCTTA | 0.000377926 |
| 229 | TBP3 | 6449 | 713 | 11.05597767 | AAAATTAAACCCTAATTTT | 0.000691105 |
| 230 | TRP1 | 1042 | 51 | 4.894433781 | AAACCCTAAACCCTAAACCCTAA | 0.00382124 |
| 231 | TRP2 | 3036 | 224 | 7.378129117 | AAACCCTAAACCCTAAACCC | 0.0039701 |
| 232 | ATY13 | 19568 | 5681 | 29.03209321 | CAACTACCAACTACC | 0.000169749 |
| 233 | BOS1 | 2175 | 806 | 37.05747126 | TTGGTAAAGTTAGGT | 0.00106963 |
| 234 | MS188 | 17081 | 6793 | 39.76933435 | AAAAATTAGGTGAAA | 1.35E-05 |
| 235 | MYB101 | 10887 | 3150 | 28.93359052 | AACCGTAACCGAATT | 0.00126249 |
| 236 | MYB107 | 13462 | 1983 | 14.7303521 | AAGGTAGGTGAACTAATT | 0.00294184 |
| 237 | MYB116 | 8961 | 3163 | 35.29739984 | AAAAAAAGTTAGGCA | 4.32E-05 |
| 238 | MYB118 | 4304 | 1639 | 38.08085502 | AAAATAACCGTTACAAA | 0.000381667 |
| 239 | MYB119 | 15557 | 13714 | 88.15324291 | TAACCGTTACA | 7.84E-05 |
| 240 | MYB121 | 2067 | 1310 | 63.3768747 | AAAGTTAGGTATAA | 2.59E-10 |
| 241 | MYB13 | 2733 | 1024 | 37.4679839 | GTAGTGGTTGGTGAA | 0.00137827 |
| 242 | MYB27 | 8363 | 4259 | 50.92670094 | AAAAAATTAGGTAAA | 0.000172562 |
| 243 | MYB33 | 22513 | 19150 | 85.0619642 | ATAACCGAATT | 0.00147965 |
| 244 | MYB3R1 | 22319 | 6164 | 27.61772481 | AAATATGACCGTTAG | 0.00322795 |
| 245 | MYB3R4 | 1965 | 823 | 41.88295165 | AAATATAACCGTTGG | 0.00360942 |
| 246 | MYB3R5 | 2706 | 981 | 36.25277162 | AAAATATGACCGTTG | 0.00200719 |
| 247 | MYB44 | 15418 | 1001 | 6.492411467 | TTTACGGTTATTTCCGTTA | 0.000494729 |
| 248 | MYB49 | 9961 | 3277 | 32.89830338 | TTGTTCACCTACCTT | 0.00368107 |
| 249 | MYB55 | 10051 | 3918 | 38.9811959 | CCTCAACCACCACC | 0.00322307 |
| 250 | MYB56 | 6039 | 2613 | 43.2687531 | ATAAATAACGGTATT | 0.000115424 |
| 251 | MYB57 | 12402 | 7833 | 63.15916788 | TTTACCTAACTTTT | 0.0031414 |
| 252 | MYB58 | 21702 | 7416 | 34.17196572 | TGTGGTTGGTGAAAC | 0.00356017 |
| 253 | MYB61 | 17756 | 4195 | 23.62581663 | TCCACCTACCACCAC | 0.00354367 |
| 254 | MYB62 | 12953 | 4460 | 34.43217787 | AAAAAAGTTAGGTAG | 0.00125896 |
| 255 | MYB63 | 2666 | 285 | 10.69017254 | CAACTACTCCACCAACCCCCA | 0.00129943 |
| 256 | MYB65 | 12876 | 3594 | 27.91239515 | ACCGTAACCGTAA | 0.000566659 |
| 257 | MYB67 | 19668 | 658 | 3.345535896 | GTAGTGGTAATGTTAGGTGGA | 0.00246673 |
| 258 | MYB70 | 11564 | 5757 | 49.78381183 | TAACCGTAATAACC | 0.00131008 |
| 259 | MYB73 | 9380 | 8119 | 86.5565032 | TAACCGTAATA | 0.000161295 |
| 260 | MYB74 | 3061 | 1936 | 63.2473048 | TTTTACCTAACTAA | 1.23E-05 |
| 261 | MYB77 | 27263 | 5324 | 19.52829843 | CGGTTATGACGGTTA | 1.01E-05 |
| 262 | MYB81 | 3231 | 2751 | 85.14391829 | GTAACCGAATT | 0.00228697 |
| 263 | MYB83 | 14502 | 10534 | 72.63825679 | GTGGTTGGTGG | 0.00328558 |
| 264 | MYB88 | 15118 | 3114 | 20.59796269 | ACCACACGCGCCTCC | 0.000153016 |
| 265 | MYB93 | 7813 | 487 | 6.233201075 | TTGGTTAAGGTAGGTGAACAA | 0.000643739 |
| 266 | MYB96 | 1147 | 253 | 22.05754141 | TAAGGTAGTTGGTAGTTG | 0.00442152 |
| 267 | MYB98 | 5558 | 2180 | 39.22274199 | AAAATGTAACGGATA | 0.00248145 |
| 268 | MYB99 | 4060 | 1169 | 28.79310345 | TCTCACCTACCTC | 0.00043355 |
| 269 | ANAC004 | 14895 | 10701 | 71.8429003 | CTTCTTCTACAAGT | 9.56E-05 |
| 270 | ANAC005 | 7671 | 1478 | 19.26737062 | TTACTTCTTTTACAAGTTT | 0.00210544 |
| 271 | ANAC013 | 5744 | 2180 | 37.95264624 | CTTGGGGAACAAGTAA | 0.00189018 |
| 272 | ANAC016 | 15693 | 5952 | 37.92773848 | CTTGCAGAACAAGCA | 5.23E-05 |
| 273 | ANAC017 | 15802 | 1753 | 11.09353246 | AGGTTTCTTGTTGAACAAG | 0.00317645 |
| 274 | ANAC020 | 7906 | 1009 | 12.76245889 | TTGCTTGTTCTACACGTTACC | 0.000142971 |
| 275 | ANAC028 | 8161 | 2654 | 32.52052445 | CTTGTTCTACAAGTAA | 0.00179026 |
| 276 | ANAC034 | 6013 | 307 | 5.105604524 | CCTGTTTTACACGGCATCT | 9.14E-06 |
| 277 | ANAC038 | 6176 | 1259 | 20.38536269 | TTACGTGTGGAACAAGTA | 0.00461066 |
| 278 | ANAC042 | 6110 | 121 | 1.980360065 | TGCCGTGATATCTACGCTAC | 0.000747267 |
| 279 | ANAC045 | 17921 | 5077 | 28.32989231 | CTTGTAGAACAAGCAA | 0.000231482 |
| 280 | ANAC046 | 18233 | 1908 | 10.46454231 | TAACGTGTAGAACAAGTAACC | 0.00102117 |
| 281 | ANAC047 | 7691 | 2339 | 30.41217007 | AAGTTACGTGTAAAA | 0.00380635 |
| 282 | ANAC050 | 21201 | 7398 | 34.89458044 | CTTGTGTCACAAGTAA | 0.00303881 |
| 283 | ANAC053 | 13096 | 6820 | 52.07697007 | TTTCTTGTGGAACAAGT | 0.000264805 |
| 284 | ANAC055 | 7513 | 492 | 6.548649008 | TTTCAAATTTTACACGTAACT | 0.000107404 |
| 285 | ANAC057 | 10212 | 3295 | 32.26596161 | TACGTGTGGAACAAGCA | 0.00124343 |
| 286 | ANAC058 | 19850 | 4172 | 21.01763224 | CTTGTGCAACACGTAA | 0.000132182 |
| 287 | ANAC062 | 10281 | 4257 | 41.40647797 | CTTAGTGATTAAGTA | 0.000451607 |
| 288 | ANAC070 | 24684 | 2133 | 8.641225085 | GGTAACGTAAAATACAAGCAA | 0.00163935 |
| 289 | ANAC071 | 23644 | 8811 | 37.26526814 | TACTTGAAGATGAAG | 0.00327323 |
| 290 | ANAC075 | 4382 | 1606 | 36.64993154 | AGCGTGAAGAAGAAG | 0.00152265 |
| 291 | ANAC079 | 2817 | 553 | 19.63081292 | GTTACGTGTAGAACAAGTAAC | 0.00215858 |
| 292 | ANAC083 | 21927 | 2899 | 13.22114288 | TTACGTGTTTTACACGTAA | 0.000385113 |
| 293 | ANAC087 | 6399 | 1044 | 16.31504923 | TAACGTGTAGAACAAGTAA | 0.00169638 |
| 294 | ANAC092 | 10364 | 3440 | 33.19181783 | CGTGTAAAACAAGCA | 0.000373493 |
| 295 | ANAC094 | 738 | 152 | 20.59620596 | TGCCGTGATCTTCACGTT | 0.00337208 |
| 296 | ANAC096 | 13107 | 3712 | 28.32074464 | CTTGAAGATGAAGCAA | 0.0017807 |
| 297 | ANAC103 | 8702 | 2287 | 26.28131464 | GGTAACTTGCTGAACAAGTTA | 0.00368258 |
| 298 | AT1G19040 | 994 | 474 | 47.6861167 | TAACTTGGGGAGCAAG | 0.00031986 |
| 299 | ATAF1 | 9197 | 219 | 2.381211265 | TACGTGTATAATTTGTCTTCTT | 0.00178942 |
| 300 | CUC1 | 3109 | 959 | 30.84593117 | CGTGTTGAACAAGTAA | 0.000465288 |
| 301 | CUC2 | 5942 | 943 | 15.87007742 | CGTGTGGAACAAGTAACCT | 0.000166017 |
| 302 | CUC3 | 6336 | 1239 | 19.55492424 | ACGTGTAAAACAAGCAAC | 6.69E-06 |
| 303 | NAC2 | 23673 | 10399 | 43.92768132 | CTTGTTCCACAAGAAAC | 0.00156117 |
| 304 | NAM | 7706 | 886 | 11.49753439 | TTACGTGTTTTACACGTAA | 6.83E-06 |
| 305 | NAP | 6299 | 5393 | 85.61676457 | ACACGTAACTT | 0.000763153 |
| 306 | NST1 | 24770 | 7115 | 28.72426322 | TAACGTAAAATTCAAGC | 0.00343412 |
| 307 | NTL8 | 5913 | 980 | 16.57365128 | GTTTCTTCATCTTCAAGTA | 0.00338041 |
| 308 | NTM1 | 15614 | 6375 | 40.82874344 | ACTTGTTCAATAAGT | 0.0011804 |
| 309 | NTM2 | 5202 | 2519 | 48.4236832 | TACTTCTTCAATAAG | 0.00386757 |
| 310 | SMB | 40054 | 2549 | 6.363908723 | GGTAGCGTGAAATTCAAGCAA | 0.00339869 |
| 311 | SND2 | 23978 | 7328 | 30.5613479 | CTTCTTCTTCACGCT | 0.00270754 |
| 312 | SND3 | 45691 | 10603 | 23.205883 | AGCGTGTGGAACAAG | 0.000827552 |
| 313 | VND1 | 7156 | 2346 | 32.78367803 | CTTGTATTTCACG | 2.94E-05 |
| 314 | VND2 | 16577 | 6009 | 36.24901973 | CTTGAAAAACAAGCA | 2.74E-05 |
| 315 | VND3 | 6534 | 2466 | 37.74104683 | CTTGAAAAACAAGCAA | 8.62E-05 |
| 316 | VND4 | 18184 | 5539 | 30.4608447 | CGTAAAAAACAAGCA | 0.000218421 |
| 317 | VND6 | 15899 | 1659 | 10.43461853 | TTGCTTGTATTTTACGCAACC | 0.00204448 |
| 318 | FRS9 | 7750 | 285 | 3.677419355 | CTCTCTCTCTCTCTCTCTCTC | 0.0033518 |
| 319 | AT1G24250 | 2233 | 469 | 21.0031348 | TCCCAACTACCAACTACC | 0.00182227 |
| 320 | BBX31 | 16775 | 6945 | 41.40089419 | AAAAAGTAAAAAAAA | 0.00017321 |
| 321 | RAV1 | 14740 | 4321 | 29.31478969 | CAGATAATTTCTGTTGT | 0.000376287 |
| 322 | REM19 | 40336 | 6679 | 16.55840936 | AAAAAAAAAAAAAAA | 0.00110976 |
| 323 | NLP7 | 867 | 294 | 33.9100346 | ATTTGGCTTTTCGGA | 0.00157036 |
| 324 | RKD2 | 10284 | 1156 | 11.24076235 | GTGACGTTTCATCTTCCTC | 0.00183413 |
| 325 | AT3G09735 | 7818 | 5005 | 64.01893067 | AGAAGCTTCTAGAAG | 5.36E-05 |
| 326 | SPL13 | 1179 | 1008 | 85.49618321 | ATTGTACGGAT | 0.00170742 |
| 327 | SPL14 | 2529 | 2239 | 88.533017 | TCCGTACAATT | 0.000148172 |
| 328 | SPL15 | 19981 | 10130 | 50.69816326 | AAATTGTACGGACA | 0.0038105 |
| 329 | SPL1 | 12940 | 11289 | 87.24111283 | AATTGTACGGA | 0.00178846 |
| 330 | SPL5 | 27217 | 15503 | 56.96072308 | AATTGTACGGAC | 0.00092055 |
| 331 | SPL9 | 41646 | 34817 | 83.60226672 | ATTGTACGGAT | 0.000546121 |
| 332 | SRS7 | 11330 | 131 | 1.156222418 | AAACCCTAAACCCTAAACCCTAA | 0.000891728 |
| 333 | At1g69690 | 995 | 719 | 72.26130653 | TGGTGGGCCCCAC | 0.000475405 |
| 334 | At1g72010 | 4626 | 2834 | 61.26242974 | ATGTGGGCCCCAC | 0.000304531 |
| 335 | At2g45680 | 450 | 45 | 10 | CCATTTGCCGTGGGTCCCACC | 0.00109515 |
| 336 | At5g08330 | 2915 | 1959 | 67.20411664 | TGTGGGCCCCACTT | 0.000256767 |
| 337 | TCP20 | 1167 | 140 | 11.99657241 | AAAAAAGCATGTGGGACCCAC | 0.00205348 |
| 338 | TCP24 | 3352 | 1999 | 59.63603819 | GTTGGGACCACA | 0.000432274 |
| 339 | TCP3 | 1416 | 648 | 45.76271186 | GTTGGGACCACAT | 0.0011376 |
| 340 | TCP7 | 1002 | 877 | 87.5249501 | GTGGGACCCAC | 0.00013153 |
| 341 | AT1G76880 | 491 | 122 | 24.84725051 | ACGGTAAAAAAATAAA | 0.00358316 |
| 342 | AT2G33550 | 8647 | 1329 | 15.36949231 | TAAAAAATATACCCTAAA | 0.000668669 |
| 343 | AT3G10030 | 3648 | 2774 | 76.04166667 | AGTTAACGGCGTTA | 0.00270821 |
| 344 | At3g14180 | 6618 | 6340 | 95.79933515 | TCGCCGGAAAA | 0.00315928 |
| 345 | AT3G25990 | 2682 | 396 | 14.76510067 | AACCATGGTTAAATTTTAACC | 8.87E-05 |
| 346 | AT5G05550 | 18951 | 9640 | 50.86802807 | TTTCTCCGGCGATGA | 0.00150621 |
| 347 | AT5G47660 | 12711 | 3815 | 30.01337424 | TTTTTTTTTTACCGT | 0.00159954 |
| 348 | GT1 | 519 | 192 | 36.99421965 | AATTAACCATGGTTAAAAT | 0.00195131 |
| 349 | GT2 | 17094 | 9608 | 56.20685621 | TTTTTTACCGTTTT | 0.000511803 |
| 350 | GT3a | 1318 | 483 | 36.64643399 | AACACGTGTTAAATTTG | 0.000100008 |
| 351 | GTL1 | 31474 | 20415 | 64.86306157 | ACGGTAAAAA | 0.000126333 |
| 352 | WRKY14 | 13405 | 6825 | 50.91383812 | AAAAGTCAACGAT | 0.00102443 |
| 353 | WRKY15 | 29394 | 25096 | 85.37796829 | AAAAGTCAACG | 0.00242641 |
| 354 | WRKY18 | 30301 | 25929 | 85.57143329 | AAAAGTCAACG | 0.00315634 |
| 355 | WRKY20 | 2213 | 1063 | 48.03434252 | AACGTTGACTATT | 0.00275611 |
| 356 | WRKY21 | 1537 | 1357 | 88.28887443 | AAAAGTCAACG | 0.00120499 |
| 357 | WRKY22 | 37563 | 17162 | 45.68857652 | AAAAGTCAACGAT | 0.0023584 |
| 358 | WRKY24 | 18866 | 11792 | 62.50397541 | TCGTTGACTTTTTT | 0.00153983 |
| 359 | WRKY25 | 20979 | 17828 | 84.98021831 | AAAAGTCAACG | 0.00471342 |
| 360 | WRKY27 | 23333 | 11001 | 47.1478164 | ATCGTTGACTTTT | 0.000406913 |
| 361 | WRKY28 | 13501 | 5830 | 43.18198652 | ATCGTTGACTTTT | 0.00181823 |
| 362 | WRKY29 | 18818 | 16438 | 87.35253481 | AAAAGTCAACG | 0.00029068 |
| 363 | WRKY30 | 4374 | 3669 | 83.88203018 | AAAGTCAACGC | 0.00091723 |
| 364 | WRKY31 | 3489 | 356 | 10.2034967 | GGATAAAAAAAAGTCAACG | 0.00225383 |
| 365 | WRKY33 | 5593 | 4840 | 86.53674236 | AAAAGTCAACG | 8.99E-07 |
| 366 | WRKY3 | 1287 | 1112 | 86.4024864 | AAAAGTCAACG | 0.000981399 |
| 367 | WRKY40 | 7114 | 2707 | 38.05172899 | AACGTTGACTTTTTT | 1.45E-05 |
| 368 | WRKY45 | 13329 | 9060 | 67.97209093 | CGTTGACTTTTT | 0.00191689 |
| 369 | WRKY50 | 13630 | 6424 | 47.13132795 | CTTTGACTTTTTT | 9.35E-05 |
| 370 | WRKY55 | 9034 | 5724 | 63.36063759 | AGCGTTGACTTT | 0.00349012 |
| 371 | WRKY65 | 20576 | 17810 | 86.55715397 | AAAAGTCAACG | 0.00306756 |
| 372 | WRKY6 | 5323 | 534 | 10.03193688 | CGTTGACTTTTTTTAACAT | 0.00268968 |
| 373 | WRKY70 | 8244 | 3749 | 45.47549733 | AGCGTTGACTTTT | 0.000529632 |
| 374 | WRKY71 | 9533 | 8152 | 85.51347949 | AAAAGTCAACG | 0.00035136 |
| 375 | WRKY75 | 29735 | 12510 | 42.07163276 | AAAAGTCAACGAC | 0.00333686 |
| 376 | WRKY7 | 1926 | 1220 | 63.34371755 | AGCGTTGACTTTTT | 0.0020648 |
| 377 | WRKY8 | 23621 | 9944 | 42.09813302 | AAAAAAGTCAACGAT | 0.00178456 |
| 378 | AT3G42860 | 2415 | 2164 | 89.60662526 | AAAAGTCAACG | 0.00105846 |
| 379 | ATHB23 | 49895 | 4178 | 8.373584528 | AATCTTAATTAATTAATAAATT | 0.000125525 |
| 380 | ATHB24 | 21428 | 11512 | 53.72409931 | AAATGTAATTAATTA | 0.00258547 |
| 381 | ATHB25 | 49795 | 29224 | 58.68862336 | TAATTAATTAAGTTT | 0.00124373 |
| 382 | AtHB32 | 21104 | 18318 | 86.79871114 | AATCGCATTAT | 6.63E-08 |
| 383 | ATHB33 | 34347 | 32395 | 94.31682534 | ACGTAATTAAT | 9.16E-06 |
| 384 | ATHB34 | 30751 | 18818 | 61.19475789 | TTTAATCATTAATTA | 0.00153627 |
Sheet 8: Information of experimentally derived motif from Plant Cistrome database of ampDAP-seq origin
| S.No. | TF | Total peaks | Covered peaks | Coverage (%) | Consensus | p-value |
|---|---|---|---|---|---|---|
| 1 | AT5G60130 | 19106 | 1537 | 8.044593321 | TTTTTGCTTATATTTTGCTTA | 0.00428143 |
| 2 | VRN1 | 7028 | 134 | 1.906659078 | CTGTTTTTTTTTTTTTTTT | 0.000286916 |
| 3 | ABR1 | 11691 | 1482 | 12.67641776 | CACCGCCGCCGCCATTTCCGC | 0.000169511 |
| 4 | AIL7 | 3535 | 1780 | 50.35360679 | ACGAATCCCGAG | 0.00113905 |
| 5 | AT1G01250 | 725 | 286 | 39.44827586 | TGTCGGTGGAGATGA | 0.00105236 |
| 6 | AT1G12630 | 18211 | 2974 | 16.33078908 | GATTTTGTCGGCAAAA | 0.00353854 |
| 7 | At1g19210 | 22451 | 6293 | 28.02993185 | CATCTCCACCGTCCA | 8.69E-05 |
| 8 | At1g22810 | 8437 | 2978 | 35.29690648 | CGCCACCGACACAGC | 0.00190908 |
| 9 | AT1G28160 | 10300 | 6627 | 64.33980583 | CGCCGCCGCCGCCA | 0.000148745 |
| 10 | At1g36060 | 4035 | 2096 | 51.94547708 | CCTCCACCGACCAT | 0.000214239 |
| 11 | AT1G71450 | 14277 | 5023 | 35.1824613 | CCTCCACCGTCAC | 0.00287054 |
| 12 | AT1G77200 | 27117 | 15708 | 57.92676181 | ATTGTCGGTGAA | 0.000298336 |
| 13 | At2g33710 | 18654 | 3879 | 20.79446767 | TCCGCCGCCGCCACCTCC | 0.000135495 |
| 14 | At2g44940 | 5163 | 1963 | 38.0205307 | ATTGTCGGTGGAG | 0.000858676 |
| 15 | AT3G16280 | 13312 | 6349 | 47.6938101 | CCACCGACAATTTT | 0.0042755 |
| 16 | AT3G60490 | 9000 | 1301 | 14.45555556 | AAAATTGTCGGTGGAG | 0.00159631 |
| 17 | At4g16750 | 15677 | 3082 | 19.6593736 | AGTGTCGGTGGAAATAA | 0.000967132 |
| 18 | At4g31060 | 7514 | 2649 | 35.25419217 | ATTTTGTCGGTGGAG | 0.00166144 |
| 19 | At4g32800 | 3779 | 733 | 19.39666578 | AATTTTGTCGGTGGAT | 0.00225151 |
| 20 | At5g65130 | 1992 | 369 | 18.52409639 | CCACCACCTCCACCGACAA | 0.00119354 |
| 21 | CBF1 | 18542 | 5367 | 28.94509762 | TGATGTCGGCGAAGA | 0.00489402 |
| 22 | CBF2 | 14034 | 4723 | 33.65398318 | TTATGTCGGCAAA | 4.08E-05 |
| 23 | CBF3 | 12163 | 9923 | 81.58349092 | TATGTCGGTAA | 0.00209348 |
| 24 | CBF4 | 29710 | 15862 | 53.38943117 | TGATGTCGGCAA | 0.0033877 |
| 25 | CEJ1 | 30321 | 10286 | 33.92368326 | AATCACCGACAAT | 0.00407569 |
| 26 | CRF10 | 6410 | 622 | 9.703588144 | ATAGAATCTTAGATTCT | 0.000606103 |
| 27 | CRF4 | 2482 | 1461 | 58.8638195 | TGGCGGCGGCGGCGG | 0.00279775 |
| 28 | DDF1 | 2395 | 977 | 40.79331942 | TCTTCGCCGACAT | 0.00435402 |
| 29 | DEAR2 | 23238 | 7026 | 30.23495998 | ATGTCGGTGGCGGTG | 0.00215012 |
| 30 | DEAR3 | 12720 | 4755 | 37.38207547 | ATTGTCGGTGATT | 3.00E-05 |
| 31 | DEAR5 | 2274 | 1897 | 83.42128408 | ATTGTCGGTGA | 0.00136611 |
| 32 | DREB19 | 15090 | 7447 | 49.35056329 | TTAAACCGACATTA | 6.56E-05 |
| 33 | ERF105 | 14787 | 1447 | 9.785622506 | CCGCCGCCGCCGCCGCCGCCGCCG | 0.0014362 |
| 34 | ERF10 | 4811 | 588 | 12.22199127 | CGGCGGAAATGGCGGCGGAG | 0.00179971 |
| 35 | ERF115 | 12101 | 4982 | 41.17015123 | GCGATGGCGGCGGCG | 8.21E-05 |
| 36 | ERF11 | 7672 | 929 | 12.10896767 | GGAAATGGCGGCGGCGGAGG | 0.00358116 |
| 37 | ERF13 | 9556 | 2919 | 30.54625366 | AAATGGCGGCGGAGG | 0.00165746 |
| 38 | ERF15 | 25902 | 4205 | 16.23426762 | CGCCGCCTCAGCCGCCAT | 0.00121631 |
| 39 | ERF2 | 726 | 207 | 28.51239669 | CCGCCGCCGCCTCCGCCGCCA | 0.00214145 |
| 40 | ERF3 | 3321 | 616 | 18.54862993 | TGGCGGCGGCGGCGGCGGCG | 0.000529809 |
| 41 | ERF4 | 15193 | 1328 | 8.740867505 | GGAAATGGCGGCGGAGGTGG | 0.000671546 |
| 42 | ERF5 | 846 | 320 | 37.8250591 | CCGCCGCCGCCGCCGCCGCCG | 3.45E-05 |
| 43 | ERF7 | 17694 | 1954 | 11.04329151 | ATGGCGGCGGAGGTGGCGGAG | 0.000564552 |
| 44 | ERF8 | 12829 | 2742 | 21.37345078 | AAAATGGCGGCGGAGGTG | 0.00301632 |
| 45 | ESE1 | 1512 | 378 | 25 | GGCGGCGGCGGCGGCGGCGG | 0.00263672 |
| 46 | ESE3 | 9328 | 4313 | 46.23713551 | GTGGCGGCGGCGGAG | 1.84E-05 |
| 47 | PUCHI | 2691 | 315 | 11.70568562 | GCGGCGGTGGCGGCGGCGGCGGAGG | 0.00459381 |
| 48 | Rap210 | 12600 | 7106 | 56.3968254 | TTTGTCGGCAAAAA | 0.00248056 |
| 49 | RAP211 | 16230 | 3769 | 23.2224276 | TTTTGGCCGGCTATT | 4.78E-07 |
| 50 | RAP21 | 2103 | 169 | 8.036138849 | CCTCCACCGACATTTCCACC | 0.000367036 |
| 51 | RRTF1 | 8037 | 1153 | 14.34614906 | GGAAATTGCGGCGGCGGTG | 0.00130955 |
| 52 | BPC1 | 17699 | 4634 | 26.18227018 | CTCTCTCTCTCTCTC | 0.00169638 |
| 53 | BAM8 | 4589 | 3027 | 65.96208324 | AGTTCACACGTGTGA | 0.000169749 |
| 54 | bHLH10 | 9144 | 3205 | 35.05030621 | ATTGTCGGTGGAG | 0.00351469 |
| 55 | bHLH122 | 10050 | 9277 | 92.30845771 | TGCAACTTGCA | 3.08E-05 |
| 56 | bHLH80 | 5667 | 5139 | 90.68290101 | TGCAACTTGCA | 0.00270851 |
| 57 | BIM1 | 647 | 97 | 14.99227202 | TGTCACGTGACTTTCACGTGCT | 0.00231702 |
| 58 | BIM2 | 40836 | 2174 | 5.32373396 | GCCACGTGAATGGCACGTG | 0.00133217 |
| 59 | ABI5 | 9233 | 3430 | 37.14935557 | AATGGTGACGTGGCA | 5.63E-05 |
| 60 | AREB3 | 4617 | 2231 | 48.32142084 | AATGGACACGTGGCA | 0.00133521 |
| 61 | bZIP16 | 8404 | 2967 | 35.30461685 | AATGCTGACGTGGCA | 4.59E-05 |
| 62 | bZIP18 | 14191 | 12259 | 86.38573744 | ATGACAGCTGG | 0.00411374 |
| 63 | bZIP44 | 2679 | 1150 | 42.9264651 | ATGCTGACGTGGC | 0.0041615 |
| 64 | bZIP48 | 5105 | 2091 | 40.95984329 | TGCCACGTCAGCATT | 0.00315198 |
| 65 | bZIP50 | 22131 | 13206 | 59.67195337 | CTGACGTCATCATC | 0.00135546 |
| 66 | bZIP52 | 16537 | 7940 | 48.01354538 | TGACCAGCTGTCAT | 0.001062 |
| 67 | bZIP53 | 17446 | 5174 | 29.65722802 | TATGCTGACGTGGCA | 6.93E-05 |
| 68 | GBF3 | 16034 | 5575 | 34.76986404 | AATGGTCACGTGGCA | 0.000984175 |
| 69 | GBF5 | 6575 | 2498 | 37.99239544 | AATGCTGACGTGGCA | 0.00151341 |
| 70 | GBF6 | 9894 | 2950 | 29.81605013 | AATGCTGACGTGGCA | 3.59E-06 |
| 71 | HY5 | 10140 | 3013 | 29.71400394 | AATGATGACGTGGCC | 0.00414024 |
| 72 | TGA10 | 5500 | 4175 | 75.90909091 | AATGACGTCATCAT | 8.56E-09 |
| 73 | TGA1 | 12377 | 6427 | 51.9269613 | AAGATGACGTCATCA | 0.00171806 |
| 74 | TGA2 | 16587 | 14392 | 86.76674504 | TGACGTCACCA | 0.000775193 |
| 75 | TGA3 | 3451 | 2346 | 67.98029557 | TGATGACGTCATC | 0.000252857 |
| 76 | TGA4 | 10140 | 5499 | 54.23076923 | AATGATGACGTCATC | 0.00274479 |
| 77 | TGA6 | 28254 | 12506 | 44.26275926 | AATGACGTCATCATC | 0.00136022 |
| 78 | TGA9 | 18677 | 17463 | 93.50002677 | TGATGACGTCA | 0.00400359 |
| 79 | VIP1 | 1523 | 955 | 62.70518713 | TGTACAGCTGTCAA | 4.07E-05 |
| 80 | At1g78700 | 5147 | 1658 | 32.21293958 | CCGCACGTGTGTATT | 0.00195143 |
| 81 | At4g18890 | 865 | 844 | 97.57225434 | CGCACGTGTGA | 0.00249362 |
| 82 | At4g36780 | 6039 | 3990 | 66.07054148 | TCACACGTGTGCAT | 0.00473557 |
| 83 | Adof1 | 11744 | 1584 | 13.48773842 | AAAAAAGAAAAAGTAAAAAAA | 0.00476697 |
| 84 | AT1G47655 | 7940 | 3229 | 40.6675063 | TTTTTTTCTTTTTTT | 0.00388283 |
| 85 | At1g64620 | 34001 | 29304 | 86.18570042 | CAAAAAGTAAA | 0.00092055 |
| 86 | AT1G69570 | 13762 | 1227 | 8.915855254 | AAAAAAAAAAAAGAAAAAGTG | 0.000449722 |
| 87 | AT2G28810 | 49623 | 19180 | 38.6514318 | TTTACTTTTTGTTTT | 0.00065049 |
| 88 | At3g45610 | 34597 | 14782 | 42.72624794 | TTTTTTTACTTTTTG | 0.00150134 |
| 89 | AT3G52440 | 30433 | 3289 | 10.80734729 | AATTTTACTTTTTCTTTTTTT | 0.000250228 |
| 90 | At4g38000 | 11840 | 9815 | 82.89695946 | GAAAAAGTTAA | 4.33E-05 |
| 91 | AT5G02460 | 45371 | 5234 | 11.53600317 | TTTTTACTTTTTTTTTTTT | 0.000505597 |
| 92 | AT5G66940 | 40237 | 4926 | 12.2424634 | TTTTTTTACTTTTTCTTTTTT | 0.000879391 |
| 93 | CDF3 | 22820 | 4763 | 20.87204207 | TTCACTTTTTCTTTTTTT | 0.00303881 |
| 94 | COG1 | 11354 | 5076 | 44.70671129 | TTTTTTCACTTTTTT | 0.00110335 |
| 95 | DAG2 | 24338 | 11441 | 47.00879283 | CAAAAAGTTAAAT | 0.000191573 |
| 96 | dof24 | 27058 | 10303 | 38.07746323 | AAAAACAAAAAGTAA | 5.20E-05 |
| 97 | dof43 | 19070 | 12832 | 67.2889355 | AAAAGCTACTTTTT | 0.00288857 |
| 98 | dof45 | 24304 | 9378 | 38.58624095 | AAAAAAGTAGCTTTT | 0.00096414 |
| 99 | OBP3 | 21664 | 2738 | 12.63847858 | TTTTTTTTTTTACTTTTTT | 0.000193396 |
| 100 | OBP4 | 27993 | 3500 | 12.50312578 | AAAAAACAAAAAGTAAAAA | 0.00189751 |
| 101 | GATA19 | 202 | 191 | 94.55445545 | CGGATCGGATT | 0.00152434 |
| 102 | GATA1 | 970 | 410 | 42.26804124 | GATTCAGATCTGAA | 0.00328534 |
| 103 | GATA20 | 17519 | 6056 | 34.56818312 | TCGATCGGATTAG | 0.00202496 |
| 104 | CRC | 9178 | 2 | 0.02179124 | CAAGTCATATTCGACTCCAAAACACTAACC | 0.00087988 |
| 105 | At5g66730 | 7624 | 2602 | 34.12906611 | TTTTTTGTCGTTTTCTG | 6.19E-05 |
| 106 | AtIDD11 | 3281 | 784 | 23.89515392 | TTTTTTTGTCGTTTTGTG | 0.000304104 |
| 107 | IDD5 | 9121 | 2887 | 31.65223112 | TTTTTTGTCGTTTTGTG | 0.000489474 |
| 108 | MGP | 3771 | 883 | 23.41553964 | TTTTTTTGTCGTTTTCTG | 0.000252257 |
| 109 | SGR5 | 18969 | 8391 | 44.23533133 | TTTTGTCTTTTTTTT | 0.00252555 |
| 110 | STOP1 | 1356 | 492 | 36.28318584 | ACCTTCCCCAGATAA | 0.0011376 |
| 111 | STZ | 36348 | 36070 | 99.23517112 | ACTTCACT | 0.000451607 |
| 112 | WIP5 | 8396 | 2451 | 29.19247261 | TACCTGGAGAACATA | 0.00398554 |
| 113 | AT3G12130 | 12290 | 8058 | 65.56550041 | AAACAAAAAGTTAA | 0.00248914 |
| 114 | AT5G63260 | 18676 | 16680 | 89.31248661 | CAAAAAGTAAA | 0.000192676 |
| 115 | CDM1 | 4777 | 2228 | 46.64015072 | CCGAAAAAATCGGAA | 0.0001645 |
| 116 | AT2G20110 | 30396 | 13188 | 43.3872878 | AATTTGAATTTTAAA | 0.000279898 |
| 117 | SOL1 | 21836 | 10313 | 47.22934603 | AATTTAAATTTTAAA | 0.000231482 |
| 118 | TCX2 | 24443 | 16938 | 69.29591294 | TTTAAAATTTAAAT | 0.000708624 |
| 119 | DEL2 | 1401 | 417 | 29.76445396 | TTTTTGGCGGGAAAA | 3.87E-05 |
| 120 | E2FA | 7135 | 3722 | 52.16538192 | TTTTGGCGCCATTT | 0.000712653 |
| 121 | FAR1 | 957 | 317 | 33.12434692 | AGGCGCGTGAAATTCACGCGC | 0.000122798 |
| 122 | At1g25550 | 15049 | 9982 | 66.3299887 | GAATAGAAGATTCC | 0.00275618 |
| 123 | AT1G49560 | 20976 | 12459 | 59.39645309 | AGAATCAAAGATTCT | 0.00124369 |
| 124 | At2g01060 | 21429 | 11055 | 51.58896822 | AAAAAAGAATATTCT | 0.00357505 |
| 125 | At2g03500 | 12731 | 6448 | 50.64802451 | GAATCTTGGATTCC | 0.00133395 |
| 126 | AT2G20400 | 6113 | 3471 | 56.78063144 | AAAGAATATTCCAAT | 0.0015998 |
| 127 | AT2G40260 | 21916 | 1102 | 5.028289834 | AAAAATTAAAAACATTCTTT | 6.64E-05 |
| 128 | At3g04030 | 22876 | 10529 | 46.02640322 | AGAATATTCCTTTTT | 0.000311417 |
| 129 | At3g12730 | 10750 | 7812 | 72.66976744 | AAAGGAATATTCTT | 0.00325747 |
| 130 | At3g24120 | 52038 | 36119 | 69.4088935 | AAGAATATTCCTTT | 0.000104312 |
| 131 | AT4G37180 | 19680 | 18245 | 92.70833333 | AAAAAGATTCC | 4.17E-06 |
| 132 | AT5G45580 | 42093 | 27809 | 66.06561661 | AAAAGGAATCTTCT | 0.00260346 |
| 133 | GRF9 | 2374 | 2352 | 99.07329402 | TGTCAGAA | 2.40E-06 |
| 134 | ANL2 | 8312 | 6573 | 79.07844081 | GCATTAATTGC | 0.00145405 |
| 135 | ATHB21 | 22139 | 1284 | 5.799719951 | ACACCAATAATTGAATATTA | 3.44E-06 |
| 136 | ATHB40 | 51300 | 28569 | 55.69005848 | ACACCAATAATTGG | 0.000387276 |
| 137 | ATHB53 | 17471 | 5366 | 30.71375422 | TCCAATTATTGGTGC | 7.73E-05 |
| 138 | ATHB5 | 46401 | 37538 | 80.89911855 | CAATGATTGA | 0.00187978 |
| 139 | LMI1 | 41115 | 19943 | 48.50541165 | TCAATTATTGGTT | 0.00317717 |
| 140 | ATHB13 | 23232 | 21189 | 91.20609504 | TCAATAATTGA | 0.000755042 |
| 141 | ATHB18 | 5366 | 4938 | 92.02385389 | CCAATGATTGG | 0.00178851 |
| 142 | ATHB20 | 5911 | 5591 | 94.58636441 | TCAATTATTGA | 0.000149727 |
| 143 | ATHB6 | 9807 | 9457 | 96.43112063 | TCAATAATTGA | 0.000264226 |
| 144 | HAT2 | 14941 | 13515 | 90.45579278 | TTAATCATTTA | 0.000560616 |
| 145 | HDG1 | 11647 | 10291 | 88.35751696 | GTAATTAATGC | 0.00236618 |
| 146 | HSF7 | 4474 | 4032 | 90.12069736 | AGAAGCTTCTAGAA | 0.000154365 |
| 147 | HSFA6B | 34926 | 21141 | 60.53083663 | AGAAGCTTCTAGA | 0.00359075 |
| 148 | HSFB3 | 1171 | 825 | 70.45260461 | AAGCTTCTAGAAG | 0.000300171 |
| 149 | HSFC1 | 8354 | 3681 | 44.06272444 | AGCTTCTAGAAGCTTCT | 0.00352171 |
| 150 | ASL18 | 6328 | 1683 | 26.59608091 | CCGGATTTTCCGGCGA | 2.91E-06 |
| 151 | LBD13 | 3715 | 1627 | 43.79542396 | CGGCGGAAATTGCGG | 0.000273712 |
| 152 | LBD18 | 20264 | 7263 | 35.84188709 | CGGATTTTCCGGAGA | 0.0008018 |
| 153 | LBD23 | 1451 | 559 | 38.52515507 | AGCGTAAAATACGCT | 0.00494099 |
| 154 | LBD2 | 24314 | 9308 | 38.282471 | CCGAAAAAATCGGAG | 1.62E-05 |
| 155 | AGL15 | 2257 | 445 | 19.71643775 | AACTTTCCTTTTTTGGAAAGT | 0.000184236 |
| 156 | SVP | 20687 | 8027 | 38.80214628 | TTCCCAAAAAGGAAA | 0.00376559 |
| 157 | At1g19000 | 29932 | 19237 | 64.26900976 | AACCTTATCCATTT | 0.00209048 |
| 158 | At1g49010 | 28552 | 10183 | 35.66475203 | AATGTGGATAAGGTT | 0.00299813 |
| 159 | AT1G72740 | 21483 | 1458 | 6.786761625 | AGTTTTAGATTTAGGGTTTTG | 0.0038348 |
| 160 | At1g74840 | 10169 | 6344 | 62.38568197 | AACCTTATCCATTT | 0.000198454 |
| 161 | At3g09600 | 29529 | 26989 | 91.39828643 | AGATATTTTTT | 0.00224349 |
| 162 | AT3G10580 | 5180 | 1343 | 25.92664093 | CCGAAAAAATCGGAA | 0.000872697 |
| 163 | At3g11280 | 2031 | 614 | 30.2314131 | TATAATCTTATCTTTATA | 0.0027039 |
| 164 | At4g01280 | 24503 | 22171 | 90.48279802 | AGATATTTTTT | 0.000124998 |
| 165 | At5g08520 | 12645 | 10411 | 82.33293792 | CCTTATCCTTA | 0.0013331 |
| 166 | At5g47390 | 17465 | 10674 | 61.11651875 | ATATGGATAAGGTT | 0.00125135 |
| 167 | At5g52660 | 19533 | 15163 | 77.62760457 | AGATATTTTTTT | 0.000906914 |
| 168 | AT5G56840 | 31403 | 6761 | 21.52979015 | TATAACCTTATCCATA | 0.000199857 |
| 169 | At5g58900 | 18957 | 8443 | 44.53763781 | AATCTTATCTTCATA | 0.000294584 |
| 170 | AT5G61620 | 19901 | 17908 | 89.98542787 | TCTTATCCAAA | 0.000238501 |
| 171 | EPR1 | 52997 | 47464 | 89.5597864 | AAAAAATATCT | 0.00293842 |
| 172 | LCL1 | 8370 | 7437 | 88.85304659 | AGATATTTTTT | 0.00171211 |
| 173 | LHY1 | 33488 | 31591 | 94.33528428 | AGATATTTTTA | 1.06E-05 |
| 174 | RVE1 | 39879 | 33634 | 84.34012889 | AGATATTTTT | 0.00209364 |
| 175 | TBP3 | 6449 | 495 | 7.675608621 | GAATTAGGGTTTAGGTTTT | 0.000855767 |
| 176 | TRP2 | 3036 | 284 | 9.354413702 | AAACCCTAAACCCTAAACCCT | 0.00170457 |
| 177 | ATY13 | 19568 | 5577 | 28.50061325 | CAACTACCAACCACC | 2.17E-05 |
| 178 | MS188 | 17081 | 6837 | 40.02693051 | AAAAAATTAGGTGAA | 0.000550485 |
| 179 | MYB101 | 10887 | 3525 | 32.37806558 | AACTGTAACCGAATT | 0.00141629 |
| 180 | MYB107 | 13462 | 6977 | 51.82736592 | TAGTTCACCTACCT | 0.00130174 |
| 181 | MYB116 | 8961 | 3441 | 38.39973217 | AAAGTTAGGCAGA | 0.00302884 |
| 182 | MYB118 | 4304 | 1914 | 44.47026022 | AAATTGTAACGGTTA | 0.00333198 |
| 183 | MYB119 | 15557 | 6493 | 41.73683872 | AAATTGTAACGGTTA | 0.0022382 |
| 184 | MYB13 | 2733 | 483 | 17.67288694 | TCCACCAACCACAATCAT | 0.00122118 |
| 185 | MYB27 | 8363 | 2951 | 35.28638049 | TTTTACCTAATTTTTT | 0.00117477 |
| 186 | MYB3R1 | 22319 | 9882 | 44.27617725 | AATATAACCGTTA | 0.00226943 |
| 187 | MYB44 | 15418 | 13343 | 86.5417045 | AATAACCGTAA | 0.00471142 |
| 188 | MYB49 | 9961 | 5862 | 58.8495131 | AAAGTTAGGTGAAA | 0.00266458 |
| 189 | MYB55 | 10051 | 3168 | 31.51925182 | TGGTTGGTGGAGTTA | 0.00436168 |
| 190 | MYB56 | 6039 | 2649 | 43.86487829 | TAAATAACGGTATTT | 0.000510282 |
| 191 | MYB58 | 21702 | 1633 | 7.524652106 | ATTGTTGTGGTTGGTGAAACA | 0.000453621 |
| 192 | MYB61 | 17756 | 5004 | 28.18202298 | CTCCACCTACCAT | 0.0020302 |
| 193 | MYB62 | 12953 | 5161 | 39.84405157 | TCTACCTAACTTT | 0.00144001 |
| 194 | MYB65 | 12876 | 3967 | 30.80925753 | ATAACCGTAACTATA | 4.67E-05 |
| 195 | MYB74 | 3061 | 1760 | 57.49754982 | AAAGGTAGGTGAAA | 0.00174302 |
| 196 | MYB77 | 27263 | 480 | 1.760627957 | CGTTTGATACGGTTACGGTT | 0.000779834 |
| 197 | MYB81 | 3231 | 1852 | 57.31971526 | ACTGTAACCGTTTT | 0.000195622 |
| 198 | MYB83 | 14502 | 4359 | 30.05792305 | TGTAGTGGTTGGTGG | 0.000380084 |
| 199 | MYB88 | 15118 | 4505 | 29.7989152 | GGAGGAGCGTGTG | 0.00425518 |
| 200 | MYB93 | 7813 | 2283 | 29.22052989 | ATTAGTTCACCTACC | 0.00152265 |
| 201 | MYB96 | 1147 | 121 | 10.54925894 | TACACCTACCAACCACCTTCT | 0.0042998 |
| 202 | MYB99 | 4060 | 3246 | 79.95073892 | TTCACCTACCT | 0.0032594 |
| 203 | ANAC004 | 14895 | 2297 | 15.42128231 | AACTTGTTCAACAAGTTTCCT | 0.00187347 |
| 204 | ANAC013 | 5744 | 2817 | 49.04247911 | ATTACTTGGAGAACAAG | 0.000160108 |
| 205 | ANAC016 | 15693 | 6317 | 40.25361626 | CTTGGAGAACAAGTA | 0.00139926 |
| 206 | ANAC017 | 15802 | 4527 | 28.64827237 | TTACTTGGAGAACAAGG | 0.000144431 |
| 207 | ANAC038 | 6176 | 864 | 13.98963731 | GGTAACTTGTAATACAAGAAA | 0.000152761 |
| 208 | ANAC045 | 17921 | 5077 | 28.32989231 | CTTGTAGAACAAGCAA | 0.00108712 |
| 209 | ANAC046 | 18233 | 4719 | 25.88164317 | CTTGTTCCACACGTTAC | 8.31E-06 |
| 210 | ANAC050 | 21201 | 9495 | 44.78562332 | TACTTGTGTAACAAG | 0.00103755 |
| 211 | ANAC053 | 13096 | 5522 | 42.16554673 | CTTGTTCCACAAGAAA | 0.00133038 |
| 212 | ANAC057 | 10212 | 2041 | 19.98629064 | AGGTTGCTTGTAGCACAAGTA | 0.000146822 |
| 213 | ANAC058 | 19850 | 5466 | 27.53652393 | TTCTTGTTTTACAAGTAA | 0.00169813 |
| 214 | ANAC062 | 10281 | 2472 | 24.04435366 | CTTAATCACTAAGTAATC | 0.00267016 |
| 215 | ANAC070 | 24684 | 4718 | 19.11359585 | CTTGAATTTCACGCAA | 0.000949294 |
| 216 | ANAC071 | 23644 | 8886 | 37.58247335 | CTTCATCTTCAAGTA | 0.00390013 |
| 217 | ANAC079 | 2817 | 822 | 29.1799787 | CTTGTTTAACACGCAA | 0.0012293 |
| 218 | ANAC083 | 21927 | 5516 | 25.15620012 | CGTGTAAAACACGCA | 4.32E-05 |
| 219 | ANAC087 | 6399 | 813 | 12.70511017 | GGTAACGTGTGGAACAAGGAA | 0.000511803 |
| 220 | ANAC092 | 10364 | 1262 | 12.17676573 | TTACTTGTTCTACACGTAAC | 0.000735053 |
| 221 | ANAC096 | 13107 | 3402 | 25.95559625 | CTTGAACTTCAAGTAACC | 0.000112609 |
| 222 | ANAC103 | 8702 | 2935 | 33.72787865 | TTGTTCAACAAGTTAC | 0.00387569 |
| 223 | CUC1 | 3109 | 470 | 15.11740109 | AGGTAACGTGTTGAACAAG | 0.000115424 |
| 224 | CUC2 | 5942 | 1637 | 27.54964658 | TAACGTGTAAAACAAG | 0.00213229 |
| 225 | CUC3 | 6336 | 1304 | 20.58080808 | CTTGTTCTACACGTTA | 7.39E-05 |
| 226 | NAC2 | 23673 | 8111 | 34.2626621 | CTTGTTCCACAAGAAA | 0.00260717 |
| 227 | NAM | 7706 | 6618 | 85.88113159 | ACACGTAACTT | 0.000818702 |
| 228 | NAP | 6299 | 1481 | 23.51166852 | TTACGTGTATAACACGG | 1.58E-05 |
| 229 | NST1 | 24770 | 7234 | 29.20468308 | CGTGAAATTCAAGCA | 0.000336462 |
| 230 | NTM1 | 15614 | 1271 | 8.140130652 | AAAAAAAAAGAAAAAGTAAAA | 0.000748242 |
| 231 | SMB | 40054 | 6938 | 17.32161582 | CGTGAAATTCAAGCAA | 0.000335573 |
| 232 | SND2 | 23978 | 7138 | 29.76895488 | AGCGTGATGAAGAAG | 0.00035428 |
| 233 | SND3 | 45691 | 1133 | 2.479700597 | GTAACTTCTTCCTCACGCTACC | 0.00300716 |
| 234 | VND2 | 16577 | 479 | 2.889545756 | TTGCTTGTATTTCACGTTACCTT | 0.000233957 |
| 235 | VND3 | 6534 | 3119 | 47.73492501 | CTTGAAAAACAAGCAAC | 0.000853804 |
| 236 | VND4 | 18184 | 3780 | 20.7875055 | CGTAAAAAACAAGCAA | 0.00411258 |
| 237 | VND6 | 15899 | 6340 | 39.87672181 | TGCTTGTTTTTCAAG | 0.000221807 |
| 238 | FRS9 | 7750 | 4 | 0.051612903 | TTGAATATTATTGGTTTAGAATTATAAAAA | 0.00202097 |
| 239 | RAV1 | 14740 | 1185 | 8.039348711 | TTTTCAGGTGATTTCTGTTGT | 0.000737465 |
| 240 | REM19 | 40336 | 1569 | 3.889825466 | ATGAAAAAAAAAAAAAAAAAA | 0.00469905 |
| 241 | RKD2 | 10284 | 4094 | 39.80941268 | GGAAGATGAAACGTC | 0.00137509 |
| 242 | SPL15 | 19981 | 5618 | 28.11671088 | ATTTGTCCGTACAAT | 1.27E-05 |
| 243 | SPL1 | 12940 | 8711 | 67.31839258 | ATCCGTACAA | 0.000119238 |
| 244 | SPL5 | 27217 | 17844 | 65.56196495 | TTGTACGGAT | 0.000139915 |
| 245 | SPL9 | 41646 | 35112 | 84.31061807 | AATTGTACGGA | 3.34E-05 |
| 246 | SRS7 | 11330 | 27 | 0.238305384 | CCCTAAACCCTAAACCCTAAACCCTAAACC | 0.000282226 |
| 247 | At1g69690 | 995 | 719 | 72.26130653 | TGGTGGGCCCCAC | 9.74E-07 |
| 248 | At1g72010 | 4626 | 2834 | 61.26242974 | ATGTGGGCCCCAC | 0.000618003 |
| 249 | At2g45680 | 450 | 310 | 68.88888889 | GTGGGACCCAC | 0.000184388 |
| 250 | At5g08330 | 2915 | 2252 | 77.25557461 | TGTGGGCCCCAC | 0.00231141 |
| 251 | TCP20 | 1167 | 893 | 76.520994 | GTGGGGCCCACC | 0.00136592 |
| 252 | TCP24 | 3352 | 2650 | 79.05727924 | GTGGGGACCAC | 0.00284685 |
| 253 | TCP3 | 1416 | 913 | 64.47740113 | GTTGGGACCACA | 7.94E-05 |
| 254 | AT2G33550 | 8647 | 5140 | 59.44258124 | AAAAATGCCCTTAA | 5.12E-05 |
| 255 | AT3G10030 | 3648 | 2195 | 60.16995614 | AGTTAACGGCGTTAA | 4.48E-05 |
| 256 | AT3G25990 | 2682 | 369 | 13.75838926 | TTTTAACTATGGTTAAGTTT | 1.56E-06 |
| 257 | AT5G05550 | 18951 | 9580 | 50.55142209 | TATCTCCGGCGACGA | 0.000647324 |
| 258 | AT5G47660 | 12711 | 3971 | 31.2406577 | TAACAACGGTAAAAA | 0.00183183 |
| 259 | GT2 | 17094 | 14239 | 83.2982333 | ACGGTAAAAAA | 6.47E-05 |
| 260 | GTL1 | 31474 | 20415 | 64.86306157 | ACGGTAAAAA | 0.000847168 |
| 261 | WRKY14 | 13405 | 6285 | 46.88549049 | AAAAGTCAACGCT | 0.00200824 |
| 262 | WRKY15 | 29394 | 17838 | 60.68585426 | GCGTTGACTTTT | 0.000681864 |
| 263 | WRKY18 | 30301 | 11972 | 39.51024719 | CGTTGACTTTTTT | 0.00050133 |
| 264 | WRKY21 | 1537 | 857 | 55.75797007 | AGCGTTGACTTTT | 5.49E-05 |
| 265 | WRKY22 | 37563 | 17129 | 45.60072412 | ATCGTTGACTTTT | 0.00434059 |
| 266 | WRKY24 | 18866 | 8736 | 46.30552316 | ATCGTTGACTTTT | 7.35E-05 |
| 267 | WRKY25 | 20979 | 7067 | 33.68606702 | AGCGTTGACTTTTTT | 0.000427131 |
| 268 | WRKY27 | 23333 | 11020 | 47.22924613 | AAAAGTCAACGAT | 0.00247562 |
| 269 | WRKY28 | 13501 | 11511 | 85.26035109 | AAAAGTCAACG | 0.00148584 |
| 270 | WRKY29 | 18818 | 8886 | 47.22074609 | AAAAGTCAACGAT | 0.0045551 |
| 271 | WRKY30 | 4374 | 2185 | 49.95427526 | AAAAGTCAACGCT | 0.000201805 |
| 272 | WRKY31 | 3489 | 1587 | 45.48581255 | AGCGTTGACTTTT | 0.00131481 |
| 273 | WRKY40 | 7114 | 3046 | 42.8169806 | AAAAAAGTCAACG | 0.00308634 |
| 274 | WRKY45 | 13329 | 11261 | 84.48495761 | AAAAGTCAACG | 1.67E-06 |
| 275 | WRKY50 | 13630 | 11731 | 86.06749817 | AAAAGTCAAAG | 6.77E-05 |
| 276 | WRKY65 | 20576 | 12955 | 62.96170295 | GCGTTGACTTTT | 0.000432639 |
| 277 | WRKY6 | 5323 | 2046 | 38.43697163 | AGCGTTGACTTTTTT | 6.42E-06 |
| 278 | WRKY75 | 29735 | 13422 | 45.13872541 | ATCGTTGACTTTT | 2.82E-05 |
| 279 | WRKY7 | 1926 | 226 | 11.73416407 | TAAAAGAAAAAAAGTCAACGA | 0.000722211 |
| 280 | WRKY8 | 23621 | 9906 | 41.93725922 | AATCGTTGACTTTTT | 0.000409345 |
| 281 | ATHB23 | 49895 | 44749 | 89.68634132 | CTTAATTAATT | 0.00189089 |
| 282 | ATHB24 | 21428 | 12049 | 56.23016614 | AAAAATAATTAATTA | 0.0021801 |
| 283 | ATHB25 | 49795 | 43207 | 86.769756 | CGTAATTAATG | 0.000209387 |
| 284 | ATHB33 | 34347 | 17576 | 51.17186363 | AAAAACGTAATTAAT | 0.000952468 |
| 285 | ATHB34 | 30751 | 15756 | 51.23735813 | TAAAACATGATTAAT | 6.67E-05 |
Sheet 9: Detailed information of docking score of all TFs
| S.No. | TF Name | Binding Affinity with motifs (Kcal/mole): A | Binding Affinity with motifs (Kcal/mole): B | Binding Affinity with motifs (Kcal/mole): C | Binding Affinity with motifs (Kcal/mole): D | Binding Affinity with motifs (Kcal/mole): E | List of interactants between TF and motif: A | List of interactants between TF and motif: B | List of interactants between TF and motif: C | List of interactants between TF and motif: D | List of interactants between TF and motif: E |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AT5G66940 | -112.277 | -110.571 | -99.236 | -109.959 | 101.946 | ARG63-A5, ARG82-T8, ARG82-G9, ARG36-T10, ARG73-T12, ASN48-T12, GLN179-C16 | ARG63-A5, ARG82-G8, ARG82-G9, ARG36-T10, ARG73-T12, ASN48-T12, GLN179-T16 | ARG63-A4, ARG82-T8, ARG82A7, ARG36-A9 | TYR46-T4, ARG63-G6, CYS62-T7, ARG64-T7, CYS37-G10 | CYS62-T11, ARG64-T11, ARG63-T10, TYR46-T8, TYR50-A5 |
| 2 | VRN1 | -84.034 | -79.186 | -72.757 | -78.458 | -70.798 | TYR260-G3, LYS298-G4, SER300-G5, ARG185-C14, ARG183-C14, GLY184-A15 | ARG185-T4, ARG296-G15, LYS298-G15, TYR260-C16, ARG183, C14 | LYS182-T4, ARG201-C2, LYS298-A12, ARG296-A13 | LYS182-T4, ARG185-T4, HIS99T16, ARG185-A14. | ARG185-T2, LYS298-A12, HIS99-A15 |
| 3 | WRKY75 | -89.625 | -84.218 | -75.814 | -85.752 | -74.324 | ARG54-T5, TYR75-T7, ARG91-A10, ARG73-C11, ARG73-C12 | ARG91-A11, LYS100-C8, GLU118-A7, LYS49-A7, LYS52-A7 | SER46-T15, LYS52-T12, LYS21-A8 | TYR94-A2, THR93-C4, LYS100-C8, LYS78-G12 | LYS78-C12, THR93-A4, TYR94-A2, GLN58-T5 |
| 4 | CEJ1 | -84.555 | -81.412 | -71.924 | -80.581 | 69.114 | LYS28-A8, ARG27-C7, ARG19-T5, ARG44-T6, ARG19-T5, LYS16-A2, ARG19-A2, LYS14-C1 | "ARG37-C6, LYS28-A10, ARG27-A11, TRP29-A11, CYS31-T14, LYS116-A16" | ARG37-A4, ARG44-A4, LEU137-C11, ASN138-A12 | LYS28-C5, ARG25-A7, ARG44-T8, ARG44-T9, LYS16-T14 | LYS28-T9, ARG-T6 |
Sheet 2: Details of MAE calculated from ten fold random independent trials.
| S.No. | Train | Test |
|---|---|---|
| 1 | 0.036950189 | 0.036536006 |
| 2 | 0.037310807 | 0.042077019 |
| 3 | 0.030275058 | 0.039225908 |
| 4 | 0.037620571 | 0.037241475 |
| 5 | 0.03764429 | 0.037027009 |
| 6 | 0.037045455 | 0.042416925 |
| 7 | 0.037645579 | 0.037014152 |
| 8 | 0.031277389 | 0.030204649 |
| 9 | 0.031238295 | 0.030556659 |
| 10 | 0.031316198 | 0.029855642 |
Sheet 3: Hyperparameter Optimization of Transformers
| Iterations | activation | activation2 | activation3 | batch_size | dropout_rate | dropout_rate2 | epochs | embed_dim | ff_dim | learning_rate | neurons | neurons2 | num_heads | optimizer | dropout_rate inside transformer | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | selu | relu | sigmoid | 16 | 0.25 | 0.25 | 25 | 28 | 14 | 0.001 | 40 | 16 | 14 | Adam | 0.15 | 0.9541 |
| 2 | LeakyReLU | relu | tanh | 111 | 0.2149 | 0.2139 | 5 | 30 | 11 | 0.8135 | 60 | 50 | 11 | Adadelta | 0.4204 | 0.9392 |
| 3 | elu | elu | relu | 118 | 0.1414 | 0.3318 | 10 | 37 | 4 | 0.8688 | 12 | 30 | 1 | Adadelta | 0.4806 | 0.9383 |
| 4 | softsign | elu | softplus | 38 | 0.01892 | 0.2404 | 11 | 91 | 11 | 0.3937 | 63 | 52 | 13 | Adagrad | 0.4785 | 0.9358 |
| 5 | selu | softplus | sigmoid | 43 | 0.2451 | 0.09475 | 16 | 58 | 10 | 0.3679 | 46 | 22 | 9 | Adam | 0.4599 | 0.9356 |
| 6 | elu | selu | selu | 131 | 0.3735 | 0.3012 | 12 | 53 | 9 | 0.3428 | 53 | 12 | 2 | Adadelta | 0.1357 | 0.9356 |
| 7 | elu | elu | sigmoid | 74 | 0.4919 | 0.2451 | 7 | 33 | 10 | 0.13 | 11 | 31 | 7 | Adadelta | 0.0741 | 0.9351 |
| 8 | elu | relu | elu | 130 | 0.3687 | 0.2657 | 10 | 96 | 16 | 0.5201 | 44 | 37 | 4 | Adam | 0.2467 | 0.935 |
| 9 | LeakyReLU | elu | tanh | 104 | 0.1337 | 0.4988 | 14 | 70 | 14 | 0.3353 | 44 | 59 | 13 | Adadelta | 0.1414 | 0.934 |
| 10 | elu | tanh | tanh | 39 | 0.2121 | 0.3741 | 6 | 34 | 13 | 0.2947 | 43 | 16 | 6 | Adadelta | 0.2129 | 0.9326 |
| 11 | relu | tanh | LeakyReLU | 110 | 0.4547 | 0.2324 | 7 | 44 | 6 | 0.688 | 19 | 19 | 13 | Adam | 0.2838 | 0.9325 |
| 12 | LeakyReLU | sigmoid | sigmoid | 103 | 0.3189 | 0.1586 | 3 | 75 | 7 | 0.7765 | 56 | 45 | 11 | Adadelta | 0.2164 | 0.9311 |
| 13 | relu | elu | softplus | 87 | 0.1418 | 0.02927 | 6 | 68 | 11 | 0.1294 | 31 | 43 | 12 | Adagrad | 0.2941 | 0.9292 |
| 14 | sigmoid | selu | sigmoid | 139 | 0.01366 | 0.4026 | 11 | 88 | 9 | 0.3873 | 30 | 22 | 11 | Adadelta | 0.1961 | 0.9284 |
| 15 | relu | sigmoid | softplus | 62 | 0.3687 | 0.04102 | 13 | 63 | 7 | 0.4923 | 8 | 52 | 10 | Adadelta | 0.4698 | 0.9276 |
| 16 | softplus | selu | softplus | 103 | 0.3099 | 0.08028 | 14 | 61 | 11 | 0.339 | 40 | 31 | 4 | Adadelta | 0.06772 | 0.9272 |
| 17 | sigmoid | selu | sigmoid | 42 | 0.2129 | 0.06985 | 6 | 72 | 8 | 0.01137 | 27 | 38 | 11 | Adagrad | 0.1053 | 0.9266 |
| 18 | selu | sigmoid | sigmoid | 23 | 0.319 | 0.3555 | 8 | 99 | 16 | 0.4727 | 58 | 29 | 11 | Adadelta | 0.3154 | 0.9266 |
| 19 | sigmoid | sigmoid | sigmoid | 24 | 0.0919 | 0.2073 | 10 | 54 | 6 | 0.4891 | 39 | 15 | 8 | Adagrad | 0.4081 | 0.9262 |
| 20 | sigmoid | selu | softplus | 82 | 0.4251 | 0.4951 | 8 | 86 | 12 | 0.3726 | 43 | 11 | 12 | Adadelta | 0.4456 | 0.9239 |
| 21 | selu | softplus | tanh | 55 | 0.2399 | 0.09335 | 9 | 65 | 15 | 0.02293 | 31 | 51 | 12 | Adamax | 0.01892 | 0.9233 |
| 22 | elu | LeakyReLU | sigmoid | 60 | 0.1633 | 0.000167 | 5 | 82 | 10 | 0.9297 | 56 | 17 | 9 | Adadelta | 0.09844 | 0.923 |
| 23 | LeakyReLU | elu | LeakyReLU | 112 | 0.1494 | 0.3173 | 3 | 27 | 11 | 0.3778 | 62 | 33 | 13 | Adadelta | 0.3702 | 0.9221 |
| 24 | sigmoid | softplus | elu | 56 | 0.368 | 0.2866 | 4 | 63 | 2 | 0.7815 | 23 | 18 | 12 | Adadelta | 0.2496 | 0.9217 |
| 25 | softplus | LeakyReLU | LeakyReLU | 87 | 0.313 | 0.4605 | 4 | 63 | 10 | 0.01558 | 43 | 43 | 3 | Adamax | 0.2445 | 0.9207 |
| 26 | tanh | selu | sigmoid | 92 | 0.1398 | 0.3254 | 15 | 90 | 12 | 0.07743 | 29 | 22 | 4 | Adagrad | 0.1402 | 0.9196 |
| 27 | sigmoid | tanh | sigmoid | 107 | 0.1657 | 0.1384 | 11 | 66 | 11 | 0.4398 | 24 | 25 | 6 | Adagrad | 0.006059 | 0.9185 |
| 28 | tanh | sigmoid | tanh | 59 | 0.3716 | 0.4354 | 5 | 51 | 8 | 0.2968 | 57 | 14 | 1 | Adadelta | 0.3849 | 0.9178 |
| 29 | softsign | softsign | sigmoid | 106 | 0.3985 | 0.02031 | 8 | 85 | 7 | 0.8161 | 20 | 31 | 14 | Adagrad | 0.04309 | 0.9156 |
| 30 | softsign | relu | sigmoid | 107 | 0.04795 | 0.2061 | 13 | 27 | 9 | 0.3129 | 13 | 46 | 3 | Adagrad | 0.4687 | 0.9143 |
| 31 | selu | elu | elu | 76 | 0.2324 | 0.3675 | 14 | 28 | 11 | 0.06012 | 46 | 36 | 7 | Adadelta | 0.2048 | 0.9141 |
| 32 | relu | relu | tanh | 94 | 0.418 | 0.1634 | 12 | 6 | 8 | 0.4165 | 45 | 34 | 10 | Adadelta | 0.3421 | 0.9135 |
| 33 | softsign | softplus | selu | 120 | 0.4966 | 0.1917 | 5 | 83 | 7 | 0.7162 | 25 | 60 | 12 | Adadelta | 0.352 | 0.9134 |
| 34 | tanh | sigmoid | tanh | 117 | 0.3375 | 0.3828 | 8 | 25 | 11 | 0.902 | 18 | 9 | 5 | Adadelta | 0.3143 | 0.9118 |
| 35 | selu | sigmoid | softplus | 94 | 0.4637 | 0.395 | 5 | 35 | 12 | 0.3756 | 58 | 34 | 5 | Adadelta | 0.4256 | 0.9106 |
| 36 | softplus | LeakyReLU | selu | 122 | 0.3336 | 0.07604 | 9 | 82 | 12 | 0.2448 | 62 | 32 | 13 | Adadelta | 0.05269 | 0.9105 |
| 37 | softplus | sigmoid | sigmoid | 111 | 0.03797 | 0.01689 | 9 | 88 | 13 | 0.1392 | 41 | 31 | 6 | Adamax | 0.1692 | 0.9092 |
| 38 | elu | softsign | tanh | 111 | 0.4408 | 0.4206 | 6 | 69 | 4 | 0.3264 | 53 | 24 | 3 | Adadelta | 0.1595 | 0.9083 |
| 39 | sigmoid | sigmoid | softsign | 124 | 0.07678 | 0.4183 | 14 | 58 | 7 | 0.2177 | 41 | 48 | 2 | Adadelta | 0.2863 | 0.9081 |
| 40 | softplus | selu | elu | 110 | 0.0977 | 0.3694 | 13 | 32 | 13 | 0.2604 | 56 | 40 | 5 | Adadelta | 0.1418 | 0.9075 |
| 41 | tanh | softsign | sigmoid | 133 | 0.2877 | 0.1626 | 7 | 79 | 11 | 0.7739 | 36 | 36 | 16 | Adagrad | 0.3687 | 0.9068 |
| 42 | elu | elu | sigmoid | 62 | 0.205 | 0.01681 | 12 | 67 | 13 | 0.1995 | 52 | 38 | 15 | Adam | 0.2381 | 0.906 |
| 43 | relu | softplus | softsign | 55 | 0.1947 | 0.4937 | 8 | 63 | 13 | 0.1754 | 34 | 50 | 11 | Adamax | 0.3364 | 0.9058 |
| 44 | sigmoid | softplus | LeakyReLU | 75 | 0.4956 | 0.0603 | 5 | 49 | 14 | 0.4854 | 22 | 32 | 7 | Adadelta | 0.4806 | 0.9054 |
| 45 | softplus | relu | softsign | 90 | 0.3991 | 0.4234 | 15 | 36 | 5 | 0.3049 | 17 | 59 | 15 | Adadelta | 0.2924 | 0.9053 |
| 46 | tanh | softsign | sigmoid | 133 | 0.1295 | 0.1214 | 15 | 66 | 14 | 0.1563 | 20 | 9 | 9 | Adamax | 0.0711 | 0.9051 |
| 47 | softplus | softplus | softplus | 105 | 0.1837 | 0.2149 | 9 | 51 | 6 | 0.135 | 18 | 42 | 14 | Adadelta | 0.2296 | 0.9023 |
| 48 | softplus | LeakyReLU | softplus | 93 | 0.3338 | 0.3027 | 11 | 62 | 11 | 0.8008 | 20 | 49 | 13 | Adadelta | 0.4308 | 0.9016 |
| 49 | elu | tanh | LeakyReLU | 29 | 0.1237 | 0.4208 | 9 | 23 | 6 | 0.02632 | 33 | 26 | 6 | Adadelta | 0.3612 | 0.9002 |
| 50 | elu | selu | sigmoid | 23 | 0.1284 | 0.07444 | 11 | 30 | 13 | 0.2176 | 62 | 38 | 2 | Adamax | 0.064 | 0.899 |
| 51 | relu | selu | tanh | 110 | 0.3181 | 0.3434 | 11 | 84 | 2 | 0.4663 | 64 | 43 | 2 | Adadelta | 0.4096 | 0.8965 |
| 52 | softsign | softsign | sigmoid | 26 | 0.39 | 0.1001 | 3 | 21 | 5 | 0.01788 | 32 | 19 | 11 | Adadelta | 0.418 | 0.8907 |
| 53 | selu | selu | LeakyReLU | 76 | 0.0177 | 0.1945 | 3 | 9 | 9 | 0.6874 | 44 | 45 | 13 | Adadelta | 0.1727 | 0.8906 |
| 54 | sigmoid | relu | elu | 134 | 0.4747 | 0.2007 | 2 | 72 | 5 | 0.3238 | 29 | 24 | 7 | Adadelta | 0.2064 | 0.8902 |
| 55 | elu | softsign | relu | 52 | 0.2547 | 0.1913 | 11 | 49 | 13 | 0.6973 | 26 | 46 | 5 | Ftrl | 0.1482 | 0.8886 |
| 56 | selu | elu | sigmoid | 61 | 0.007987 | 0.02023 | 11 | 86 | 8 | 0.3065 | 18 | 15 | 8 | Adamax | 0.04266 | 0.8789 |
| 57 | LeakyReLU | softsign | selu | 36 | 0.2149 | 0.003288 | 7 | 47 | 4 | 0.7053 | 27 | 18 | 16 | Ftrl | 0.4305 | 0.8742 |
| 58 | elu | LeakyReLU | softsign | 90 | 0.1098 | 0.194 | 13 | 36 | 2 | 0.02138 | 52 | 29 | 4 | Adadelta | 0.0974 | 0.8714 |
| 59 | sigmoid | softsign | LeakyReLU | 79 | 0.1917 | 0.3749 | 9 | 99 | 15 | 0.119 | 20 | 48 | 3 | Adamax | 0.4066 | 0.8289 |
| 60 | selu | softplus | sigmoid | 53 | 0.1055 | 0.4211 | 13 | 43 | 6 | 0.2092 | 35 | 12 | 13 | Adamax | 0.246 | 0.8247 |
| 61 | sigmoid | softplus | softsign | 57 | 0.2496 | 0.01173 | 13 | 92 | 11 | 0.02486 | 30 | 28 | 14 | Adadelta | 0.4573 | 0.8153 |
| 62 | softplus | sigmoid | LeakyReLU | 95 | 0.3783 | 0.08313 | 15 | 87 | 9 | 0.01262 | 20 | 35 | 16 | Adagrad | 0.01366 | 0.8088 |
| 63 | LeakyReLU | LeakyReLU | LeakyReLU | 105 | 0.2064 | 0.1528 | 12 | 42 | 5 | 0.5889 | 45 | 42 | 3 | Ftrl | 0.2073 | 0.7934 |
| 64 | sigmoid | sigmoid | sigmoid | 106 | 0.2705 | 0.3486 | 7 | 37 | 8 | 0.01822 | 46 | 8 | 16 | Adagrad | 0.186 | 0.7908 |
| 65 | softsign | tanh | tanh | 125 | 0.2617 | 0.4043 | 13 | 46 | 3 | 0.8458 | 48 | 61 | 3 | Ftrl | 0.399 | 0.7711 |
| 66 | tanh | tanh | softsign | 28 | 0.2537 | 0.2679 | 6 | 21 | 6 | 0.01 | 33 | 23 | 8 | Adadelta | 0.1932 | 0.7695 |
| 67 | relu | softplus | sigmoid | 89 | 0.2118 | 0.1496 | 10 | 33 | 4 | 0.4032 | 46 | 9 | 15 | Adam | 0.009722 | 0.7675 |
| 68 | LeakyReLU | sigmoid | selu | 75 | 0.3489 | 0.07585 | 11 | 91 | 16 | 0.01964 | 46 | 22 | 4 | Adagrad | 0.03249 | 0.7674 |
| 69 | selu | softsign | softsign | 46 | 0.006059 | 0.01772 | 7 | 61 | 14 | 0.08508 | 20 | 38 | 6 | Adadelta | 0.4145 | 0.751 |
| 70 | sigmoid | tanh | softsign | 117 | 0.352 | 0.2095 | 5 | 30 | 3 | 0.2429 | 29 | 51 | 6 | Adamax | 0.4251 | 0.7408 |
| 71 | softsign | softsign | tanh | 114 | 0.325 | 0.1155 | 10 | 59 | 7 | 0.8352 | 54 | 37 | 8 | Ftrl | 0.3048 | 0.7381 |
| 72 | tanh | softplus | LeakyReLU | 56 | 0.392 | 0.007872 | 10 | 78 | 13 | 0.03935 | 49 | 12 | 12 | Adagrad | 0.2358 | 0.7348 |
| 73 | sigmoid | softplus | sigmoid | 46 | 0.04452 | 0.04207 | 13 | 98 | 9 | 0.4093 | 10 | 36 | 9 | Adamax | 0.07834 | 0.7328 |
| 74 | elu | selu | softsign | 71 | 0.1497 | 0.3439 | 4 | 62 | 11 | 0.4853 | 58 | 29 | 15 | Ftrl | 0.1055 | 0.7214 |
| 75 | softsign | sigmoid | sigmoid | 61 | 0.3704 | 0.4621 | 4 | 51 | 3 | 0.02806 | 40 | 16 | 13 | Nadam | 0.2118 | 0.7152 |
| 76 | sigmoid | selu | tanh | 59 | 0.4308 | 0.07067 | 13 | 53 | 6 | 0.8311 | 46 | 54 | 4 | Adadelta | 0.01205 | 0.6704 |
| 77 | relu | sigmoid | sigmoid | 33 | 0.402 | 0.157 | 11 | 86 | 14 | 0.6356 | 22 | 9 | 11 | Adagrad | 0.319 | 0.6704 |
| 78 | sigmoid | elu | sigmoid | 123 | 0.4244 | 0.244 | 13 | 78 | 16 | 0.7103 | 45 | 18 | 7 | Adagrad | 0.3133 | 0.6584 |
| 79 | sigmoid | tanh | softplus | 129 | 0.2924 | 0.2615 | 12 | 74 | 9 | 0.29 | 37 | 31 | 5 | Adamax | 0.2402 | 0.6467 |
| 80 | tanh | softsign | softplus | 94 | 0.3517 | 0.03177 | 10 | 50 | 9 | 0.344 | 35 | 21 | 12 | Adamax | 0.2561 | 0.6438 |
| 81 | tanh | softplus | softsign | 44 | 0.06839 | 0.1863 | 13 | 13 | 11 | 0.3004 | 26 | 11 | 10 | Adagrad | 0.1845 | 0.6424 |
| 82 | relu | softsign | softsign | 74 | 0.1038 | 0.1851 | 8 | 28 | 6 | 0.2274 | 22 | 21 | 10 | Adagrad | 0.02065 | 0.6338 |
| 83 | relu | selu | elu | 127 | 0.4305 | 0.1761 | 11 | 15 | 3 | 0.04056 | 51 | 15 | 15 | Adamax | 0.2222 | 0.6255 |
| 84 | tanh | selu | sigmoid | 100 | 0.448 | 0.08714 | 6 | 8 | 11 | 0.5194 | 17 | 44 | 8 | Adam | 0.0596 | 0.6147 |
| 85 | softsign | relu | sigmoid | 69 | 0.4962 | 0.02624 | 11 | 75 | 16 | 0.7116 | 8 | 21 | 16 | Adagrad | 0.4584 | 0.5841 |
| 86 | softplus | sigmoid | softplus | 88 | 0.03063 | 0.1134 | 11 | 70 | 7 | 0.06583 | 60 | 27 | 7 | Adagrad | 0.3071 | 0.5832 |
| 87 | LeakyReLU | LeakyReLU | sigmoid | 107 | 0.05785 | 0.2679 | 7 | 14 | 10 | 0.764 | 55 | 10 | 13 | Adamax | 0.05785 | 0.5815 |
| 88 | softsign | sigmoid | tanh | 106 | 0.4806 | 0.4375 | 3 | 42 | 3 | 0.3595 | 40 | 50 | 12 | Adamax | 0.4904 | 0.5802 |
| 89 | sigmoid | relu | softsign | 125 | 0.4049 | 0.3307 | 11 | 94 | 13 | 0.1461 | 36 | 28 | 8 | Adam | 0.225 | 0.5702 |
| 90 | selu | LeakyReLU | sigmoid | 40 | 0.04818 | 0.2455 | 3 | 34 | 15 | 0.8042 | 9 | 31 | 16 | Nadam | 0.3716 | 0.5524 |
| 91 | elu | tanh | softsign | 47 | 0.4526 | 0.4392 | 15 | 16 | 10 | 0.02637 | 62 | 44 | 15 | Ftrl | 0.4966 | 0.5473 |
| 92 | softsign | elu | elu | 62 | 0.3584 | 0.2945 | 2 | 65 | 11 | 0.1297 | 14 | 21 | 2 | Ftrl | 0.2285 | 0.5472 |
| 93 | softplus | sigmoid | relu | 95 | 0.0596 | 0.2751 | 3 | 53 | 8 | 0.0141 | 16 | 30 | 2 | SGD | 0.1191 | 0.5274 |
| 94 | LeakyReLU | softsign | sigmoid | 92 | 0.3059 | 0.2436 | 3 | 10 | 10 | 0.7463 | 38 | 39 | 1 | Adamax | 0.285 | 0.5267 |
| 95 | tanh | tanh | elu | 92 | 0.4066 | 0.1238 | 6 | 6 | 14 | 0.2815 | 29 | 34 | 3 | Ftrl | 0.003066 | 0.5265 |
| 96 | sigmoid | tanh | softplus | 99 | 0.02393 | 0.02245 | 5 | 96 | 12 | 0.03158 | 46 | 36 | 7 | Adam | 0.448 | 0.5009 |
| 97 | LeakyReLU | selu | selu | 122 | 0.4698 | 0.000829 | 4 | 93 | 7 | 0.8852 | 38 | 13 | 1 | Nadam | 0.4702 | 0.5009 |
| 98 | elu | LeakyReLU | softplus | 87 | 0.4599 | 0.3602 | 10 | 44 | 13 | 0.563 | 34 | 18 | 2 | Ftrl | 0.01635 | 0.5007 |
| 99 | softplus | softplus | relu | 66 | 0.003241 | 0.2312 | 15 | 73 | 5 | 0.1078 | 51 | 24 | 10 | SGD | 0.3019 | 0.5001 |
| 100 | tanh | elu | relu | 42 | 0.1482 | 0.193 | 10 | 40 | 15 | 0.3548 | 18 | 12 | 7 | Adamax | 0.4633 | 0.5001 |
| 101 | LeakyReLU | sigmoid | selu | 130 | 0.4584 | 0.0463 | 8 | 91 | 16 | 0.3947 | 16 | 39 | 11 | RMSprop | 0.0116 | 0.5001 |
| 102 | relu | LeakyReLU | elu | 79 | 0.002124 | 0.4942 | 14 | 78 | 16 | 0.6762 | 64 | 34 | 6 | Adam | 0.1497 | 0.5001 |
| 103 | relu | sigmoid | sigmoid | 27 | 0.2048 | 0.2185 | 15 | 55 | 15 | 0.4083 | 17 | 55 | 16 | Adamax | 0.2617 | 0.5001 |
| 104 | tanh | relu | LeakyReLU | 108 | 0.07831 | 0.008304 | 12 | 35 | 14 | 0.6245 | 18 | 57 | 11 | SGD | 0.1337 | 0.5001 |
| 105 | softplus | softsign | sigmoid | 132 | 0.03249 | 0.3181 | 6 | 38 | 12 | 0.1263 | 57 | 52 | 11 | RMSprop | 0.02454 | 0.5001 |
| 106 | softplus | sigmoid | relu | 40 | 0.2421 | 0.2786 | 3 | 28 | 2 | 0.7352 | 27 | 43 | 4 | Nadam | 0.4288 | 0.5001 |
| 107 | softplus | elu | relu | 20 | 0.4785 | 0.3873 | 7 | 96 | 7 | 0.786 | 49 | 61 | 11 | Nadam | 0.2075 | 0.5001 |
| 108 | softplus | sigmoid | sigmoid | 108 | 0.3767 | 0.4396 | 9 | 82 | 14 | 0.8111 | 31 | 59 | 12 | Nadam | 0.2815 | 0.5001 |
| 109 | sigmoid | softsign | softsign | 106 | 0.399 | 0.1984 | 7 | 80 | 4 | 0.6715 | 28 | 34 | 5 | Adam | 0.07896 | 0.5001 |
| 110 | LeakyReLU | LeakyReLU | LeakyReLU | 83 | 0.09375 | 0.2922 | 6 | 62 | 15 | 0.9075 | 39 | 31 | 15 | Adamax | 0.01655 | 0.5001 |
| 111 | relu | tanh | elu | 27 | 0.4604 | 0.2097 | 16 | 65 | 5 | 0.3287 | 54 | 31 | 11 | SGD | 0.2866 | 0.5001 |
| 112 | sigmoid | LeakyReLU | relu | 23 | 0.01635 | 0.05767 | 10 | 30 | 5 | 0.2794 | 15 | 17 | 13 | RMSprop | 0.2298 | 0.5001 |
| 113 | tanh | selu | sigmoid | 32 | 0.2544 | 0.21 | 14 | 64 | 5 | 0.4299 | 10 | 14 | 4 | SGD | 0.3517 | 0.5001 |
| 114 | elu | softplus | elu | 113 | 0.3612 | 0.1378 | 16 | 97 | 5 | 0.8679 | 35 | 22 | 8 | Adagrad | 0.4604 | 0.5001 |
| 115 | softplus | relu | elu | 63 | 0.2684 | 0.18 | 3 | 75 | 6 | 0.884 | 13 | 57 | 7 | Adamax | 0.4506 | 0.5001 |
| 116 | selu | softsign | LeakyReLU | 115 | 0.02155 | 0.3345 | 15 | 59 | 9 | 0.2079 | 14 | 40 | 12 | Adagrad | 0.1504 | 0.5001 |
| 117 | sigmoid | selu | relu | 65 | 0.4735 | 0.1846 | 2 | 39 | 9 | 0.5956 | 42 | 51 | 12 | Nadam | 0.3242 | 0.5001 |
| 118 | softsign | LeakyReLU | relu | 68 | 0.064 | 0.2827 | 15 | 97 | 7 | 0.6654 | 42 | 43 | 13 | Nadam | 0.0919 | 0.5001 |
| 119 | softsign | LeakyReLU | relu | 108 | 0.1727 | 0.07877 | 16 | 11 | 5 | 0.8693 | 34 | 35 | 14 | Adamax | 0.4319 | 0.5001 |
| 120 | elu | softsign | softplus | 74 | 0.4299 | 0.09439 | 10 | 38 | 10 | 0.2112 | 29 | 22 | 14 | Adagrad | 0.4238 | 0.5001 |
| 121 | sigmoid | relu | LeakyReLU | 23 | 0.2793 | 0.2825 | 14 | 23 | 14 | 0.996 | 60 | 50 | 10 | Adamax | 0.435 | 0.5001 |
| 122 | softsign | LeakyReLU | selu | 77 | 0.3933 | 0.3014 | 13 | 11 | 15 | 0.5458 | 21 | 44 | 11 | Nadam | 0.1917 | 0.5001 |
| 123 | softplus | sigmoid | selu | 115 | 0.4633 | 0.1839 | 2 | 54 | 13 | 0.5886 | 49 | 18 | 6 | Adagrad | 0.0839 | 0.5001 |
| 124 | softsign | relu | relu | 117 | 0.2537 | 0.4487 | 13 | 24 | 13 | 0.4284 | 29 | 38 | 7 | Adamax | 0.3707 | 0.5001 |
| 125 | LeakyReLU | tanh | relu | 30 | 0.296 | 0.05701 | 13 | 10 | 13 | 0.3856 | 37 | 14 | 4 | Nadam | 0.4683 | 0.5001 |
| 126 | sigmoid | relu | sigmoid | 134 | 0.186 | 0.444 | 13 | 52 | 11 | 0.1223 | 20 | 53 | 10 | Nadam | 0.394 | 0.5001 |
| 127 | selu | elu | LeakyReLU | 92 | 0.01205 | 0.3945 | 12 | 36 | 9 | 0.6353 | 63 | 29 | 1 | Adam | 0.2278 | 0.5001 |
| 128 | softplus | sigmoid | selu | 64 | 0.2222 | 0.3026 | 6 | 46 | 7 | 0.9235 | 46 | 15 | 7 | Adagrad | 0.2408 | 0.5001 |
| 129 | tanh | selu | sigmoid | 21 | 0.4386 | 0.3638 | 13 | 24 | 4 | 0.3859 | 40 | 51 | 5 | Adamax | 0.2877 | 0.5001 |
| 130 | sigmoid | softplus | relu | 99 | 0.4081 | 0.3511 | 14 | 60 | 7 | 0.5283 | 23 | 25 | 5 | SGD | 0.3721 | 0.5001 |
| 131 | softplus | sigmoid | softplus | 125 | 0.4072 | 0.3249 | 3 | 47 | 13 | 0.6756 | 34 | 34 | 16 | RMSprop | 0.1295 | 0.5001 |
| 132 | relu | sigmoid | LeakyReLU | 116 | 0.1174 | 0.2039 | 11 | 75 | 5 | 0.3516 | 31 | 49 | 12 | SGD | 0.3735 | 0.5001 |
| 133 | relu | LeakyReLU | selu | 46 | 0.3154 | 0.316 | 4 | 11 | 5 | 0.6724 | 56 | 21 | 4 | Adamax | 0.2968 | 0.5001 |
| 134 | tanh | sigmoid | softsign | 130 | 0.266 | 0.1197 | 10 | 79 | 16 | 0.1073 | 23 | 42 | 15 | Ftrl | 0.3302 | 0.5 |
| 135 | sigmoid | softsign | relu | 56 | 0.3421 | 0.07301 | 11 | 43 | 12 | 0.3962 | 48 | 43 | 9 | Adam | 0.3667 | 0.5 |
| 136 | softsign | relu | selu | 101 | 0.02454 | 0.1969 | 15 | 89 | 15 | 0.831 | 22 | 55 | 12 | Adamax | 0.3197 | 0.5 |
| 137 | relu | sigmoid | sigmoid | 65 | 0.4319 | 0.06493 | 15 | 42 | 11 | 0.9416 | 30 | 55 | 11 | Adagrad | 0.1393 | 0.5 |
| 138 | softsign | tanh | softsign | 69 | 0.3682 | 0.1219 | 7 | 97 | 7 | 0.1986 | 35 | 51 | 4 | SGD | 0.2556 | 0.5 |
| 139 | softplus | LeakyReLU | softsign | 107 | 0.009045 | 0.1358 | 9 | 25 | 13 | 0.9759 | 21 | 29 | 15 | SGD | 0.3196 | 0.5 |
| 140 | LeakyReLU | LeakyReLU | softplus | 109 | 0.246 | 0.4638 | 15 | 18 | 14 | 0.5974 | 30 | 38 | 15 | Ftrl | 0.249 | 0.5 |
| 141 | elu | selu | tanh | 134 | 0.2358 | 0.385 | 12 | 76 | 13 | 0.5863 | 18 | 48 | 10 | RMSprop | 0.3338 | 0.5 |
| 142 | LeakyReLU | LeakyReLU | LeakyReLU | 35 | 0.2367 | 0.3989 | 8 | 58 | 14 | 0.02545 | 20 | 38 | 2 | RMSprop | 0.02393 | 0.5 |
| 143 | elu | softplus | tanh | 83 | 0.2296 | 0.4658 | 9 | 22 | 11 | 0.4022 | 49 | 53 | 6 | Adagrad | 0.01781 | 0.5 |
| 144 | relu | relu | tanh | 67 | 0.2143 | 0.2768 | 13 | 92 | 12 | 0.04397 | 25 | 11 | 3 | Nadam | 0.3675 | 0.5 |
| 145 | sigmoid | tanh | softsign | 67 | 0.04273 | 0.488 | 9 | 100 | 12 | 0.5786 | 57 | 54 | 14 | Adamax | 0.2143 | 0.5 |
| 146 | selu | relu | relu | 58 | 0.2445 | 0.448 | 14 | 75 | 13 | 0.1843 | 17 | 11 | 13 | RMSprop | 0.2451 | 0.5 |
| 147 | softplus | relu | sigmoid | 68 | 0.08005 | 0.493 | 10 | 79 | 8 | 0.6739 | 52 | 8 | 2 | RMSprop | 0.4611 | 0.5 |
| 148 | sigmoid | LeakyReLU | relu | 27 | 0.2556 | 0.3757 | 9 | 49 | 2 | 0.5929 | 55 | 56 | 2 | Nadam | 0.4962 | 0.5 |
| 149 | tanh | relu | selu | 133 | 0.1393 | 0.4083 | 3 | 91 | 7 | 0.1574 | 18 | 16 | 6 | Nadam | 0.4545 | 0.5 |
| 150 | sigmoid | sigmoid | relu | 61 | 0.4872 | 0.464 | 16 | 58 | 7 | 0.6239 | 24 | 27 | 15 | Adagrad | 0.1092 | 0.5 |
Table S3: Details for nine different studies done in the MS which cover i) Impact of words representation on the transformers performance. ii) sheets 2-3 cover performance of PTFSpot on the datasets "A" and "B". iii) sheets 4-5 cover details on 10 fold independent random trials for datasets "A" and"B". iv) sheets 6-7 cover performance benchmarking studies on datasets "B" and "C". v) Sheet 8-9: Details of the study showing the variations in sequence and domains of the transcription factors when compared between A. thaliana and Z. mays.
Sheet 1: Evaluation of word representations
| S.No. | Word representation | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| 1 | Dimer (Single) | 82.1501183 | 79.4100786 | 80.7780008 |
| 2 | Pentamer (Single) | 86.5270132 | 83.5823127 | 85.0534887 |
| 3 | Heptamer (Single) | 87.077462 | 85.3997082 | 86.2382013 |
| 4 | Dimer | 85.3442593 | 86.0933061 | 85.7187827 |
| 5 | Pentamer | 87.482382 | 87.6233818 | 87.5528819 |
| 6 | Heptamer | 88.6441998 | 88.2085785 | 88.4263891 |
| 7 | Dimer + Pentamer | 88.9173543 | 87.6997151 | 88.3141625 |
| 8 | Dimer + Heptamer | 90.4255611 | 91.3351223 | 90.8803417 |
| 9 | Pentamer + Heptamer | 94.3450133 | 95.4610128 | 94.903013 |
| 10 | Dimer + Pentamer + Heptamer | 95.331545 | 95.8015201 | 95.5659375 |
Sheet 2: Performace of PTFSpot on Dataset 'A' TFs
| S.No. | Name | Sensitivity | Specificity | Accuracy | F1 Score | MCC | AUC |
|---|---|---|---|---|---|---|---|
| 1 | ABF1 | 0.934884787 | 0.945259658 | 0.940072223 | 0.93975973 | 0.88732106 | 0.940072223 |
| 2 | ABF3 | 0.902927071 | 0.924259964 | 0.913593517 | 0.912661931 | 0.842083284 | 0.913593517 |
| 3 | ABF4 | 0.932674459 | 0.945667988 | 0.939171224 | 0.938773449 | 0.885733084 | 0.939171224 |
| 4 | ABI5 | 0.953809524 | 0.974285714 | 0.964047619 | 0.963675728 | 0.930665871 | 0.964047619 |
| 5 | ANAC032 | 0.962465822 | 0.9550087 | 0.958737261 | 0.95889054 | 0.920877549 | 0.958737261 |
| 6 | ANAC102 | 0.962743091 | 0.94370522 | 0.953224156 | 0.953665213 | 0.910808122 | 0.953224156 |
| 7 | AP1 | 0.922714227 | 0.954694547 | 0.938704387 | 0.937708333 | 0.884864317 | 0.938704387 |
| 8 | AZF1 | 0.925174382 | 0.96956246 | 0.947368421 | 0.9461738 | 0.900179107 | 0.947368421 |
| 9 | BZIP28 | 0.982154238 | 0.978330147 | 0.980242192 | 0.980279898 | 0.961264844 | 0.980242192 |
| 10 | CCA1 | 0.982428115 | 0.993610224 | 0.988019169 | 0.987951807 | 0.976323941 | 0.988019169 |
| 11 | CRY2 | 0.907798363 | 0.931925894 | 0.919862128 | 0.918883559 | 0.852525525 | 0.919862128 |
| 12 | DELLA | 0.932374101 | 0.953956835 | 0.943165468 | 0.942545455 | 0.892766312 | 0.943165468 |
| 13 | DREB2A | 0.948019283 | 0.916998533 | 0.932508908 | 0.933539732 | 0.874067421 | 0.932508908 |
| 14 | FBH3 | 0.92680012 | 0.93576337 | 0.931281745 | 0.930972389 | 0.872002746 | 0.931281745 |
| 15 | FHY1 | 0.972307692 | 0.972307692 | 0.972307692 | 0.972307692 | 0.946149112 | 0.972307692 |
| 16 | FHY3 | 0.989417989 | 0.986772487 | 0.988095238 | 0.988110964 | 0.976473841 | 0.988095238 |
| 17 | FIE | 0.943030303 | 0.96 | 0.951515152 | 0.951100244 | 0.907718586 | 0.951515152 |
| 18 | FLM | 0.908176944 | 0.960455764 | 0.934316354 | 0.932553338 | 0.877094255 | 0.934316354 |
| 19 | GBF2 | 0.936043224 | 0.944273418 | 0.940158321 | 0.939911049 | 0.887474885 | 0.940158321 |
| 20 | HAT22 | 0.932623873 | 0.938114282 | 0.935369077 | 0.935191164 | 0.879090645 | 0.935369077 |
| 21 | HB5 | 0.960553163 | 0.961573339 | 0.961063251 | 0.96104338 | 0.925158604 | 0.961063251 |
| 22 | HB7 | 0.955434479 | 0.952780106 | 0.954107293 | 0.95416812 | 0.912426558 | 0.954107293 |
| 23 | HSFA1A | 0.930831333 | 0.952214707 | 0.94152302 | 0.940891045 | 0.889859997 | 0.94152302 |
| 24 | HSFA6A | 0.934551295 | 0.951616141 | 0.943083718 | 0.942593905 | 0.892630739 | 0.943083718 |
| 25 | IBH1 | 0.945985401 | 0.951824818 | 0.948905109 | 0.94875549 | 0.903029942 | 0.948905109 |
| 26 | JAG | 0.961841308 | 0.955784373 | 0.958812841 | 0.958937198 | 0.921016997 | 0.958812841 |
| 27 | KAN1 | 0.971608054 | 0.959167828 | 0.965387941 | 0.965601901 | 0.933166702 | 0.965387941 |
| 28 | LEC1 | 0.975927033 | 0.962320574 | 0.969123804 | 0.969332442 | 0.94014875 | 0.969123804 |
| 29 | LFY | 0.942889454 | 0.936952219 | 0.939920837 | 0.940098661 | 0.887058695 | 0.939920837 |
| 30 | MYB3 | 0.933911159 | 0.934607646 | 0.934259403 | 0.934236501 | 0.877162428 | 0.934259403 |
| 31 | NFYB2 | 0.92376409 | 0.95388429 | 0.93882419 | 0.937888789 | 0.885081301 | 0.93882419 |
| 32 | NFYC2 | 0.941939287 | 0.951724138 | 0.946831712 | 0.946570312 | 0.899312339 | 0.946831712 |
| 33 | PIF1 | 0.950166113 | 0.961517165 | 0.955841639 | 0.955589587 | 0.915577763 | 0.955841639 |
| 34 | PIF4 | 0.8746044 | 0.918911617 | 0.896758008 | 0.894419009 | 0.814652297 | 0.896758008 |
| 35 | PIF5 | 0.939457203 | 0.958594294 | 0.949025748 | 0.948533286 | 0.90323054 | 0.949025748 |
| 36 | REV | 0.970911441 | 0.959599224 | 0.965255333 | 0.965450747 | 0.93292076 | 0.965255333 |
| 37 | RGA | 0.961838499 | 0.953826692 | 0.957832595 | 0.95800084 | 0.919218779 | 0.957832595 |
| 38 | SOC1 | 0.94368071 | 0.963192905 | 0.953436807 | 0.952978056 | 0.911192987 | 0.953436807 |
| 39 | WRKY33 | 0.977304783 | 0.964696329 | 0.971000556 | 0.971182229 | 0.943678573 | 0.971000556 |
| 40 | ZAT6 | 0.938588885 | 0.942294048 | 0.940441466 | 0.940330924 | 0.887976602 | 0.940441466 |
| 41 | AT5G04760 | 0.997196548 | 0.998272884 | 0.99773859 | 0.997720998 | 0.995487137 | 0.997734716 |
| 42 | BBM | 0.985947794 | 0.986519505 | 0.986232188 | 0.98629737 | 0.972842863 | 0.986233649 |
| 43 | ERF115 | 0.994860132 | 0.992738495 | 0.993798133 | 0.993797851 | 0.987673177 | 0.993799313 |
| 44 | GBF3 | 0.999742931 | 0.999903599 | 0.999823265 | 0.999823251 | 0.999646592 | 0.999823265 |
| 45 | HB6 | 1 | 1 | 1 | 1 | 1 | 1 |
| 46 | MYB3R3 | 0.948737764 | 0.959531687 | 0.9541672 | 0.953650958 | 0.912522551 | 0.954134726 |
| 47 | MYB44 | 0.999712282 | 0.999964427 | 0.999839044 | 0.999838138 | 0.99967813 | 0.999838354 |
| 48 | PhyA | 0.95287187 | 0.955817378 | 0.954344624 | 0.954277286 | 0.912857698 | 0.954344624 |
| 49 | RD26 | 0.999967834 | 0.999967852 | 0.999967843 | 0.999967834 | 0.999935688 | 0.999967843 |
| 50 | SEP3 | 0.985596107 | 0.979240261 | 0.982409317 | 0.982416996 | 0.965437132 | 0.982418184 |
| 51 | SVP | 0.963632356 | 0.958 | 0.960827708 | 0.96109035 | 0.924720393 | 0.960816178 |
Sheet 3: Performace of PTFSpot on Dataset 'B' TFs
| S.No. | Name | Sensitivity | Specificity | Accuracy | F1 Score | MCC | AUC |
|---|---|---|---|---|---|---|---|
| 1 | ABF2 | 0.984658298 | 0.995475113 | 0.989855072 | 0.990182328 | 0.979893041 | 0.989855072 |
| 2 | ABI5 | 0.980733605 | 0.979061372 | 0.979886634 | 0.979644708 | 0.960576747 | 0.979886634 |
| 3 | ABR1 | 0.982300885 | 0.992596811 | 0.987455453 | 0.987374462 | 0.975223967 | 0.987455453 |
| 4 | Adof1 | 0.984414848 | 0.985503127 | 0.984958138 | 0.984973065 | 0.970368753 | 0.984958138 |
| 5 | AGL15 | 0.811614731 | 0.861325116 | 0.835424354 | 0.837107378 | 0.72471253 | 0.835424354 |
| 6 | AGL63 | 0.9094728 | 0.952411438 | 0.930771674 | 0.929783122 | 0.87104056 | 0.930771674 |
| 7 | AIL7 | 0.8633235 | 0.875482625 | 0.869459328 | 0.867588933 | 0.772949706 | 0.869459328 |
| 8 | ANAC004 | 0.911206897 | 0.935368043 | 0.924773185 | 0.913964548 | 0.858632532 | 0.924773185 |
| 9 | ANAC005 | 0.905198777 | 0.945548833 | 0.92548338 | 0.923556942 | 0.861930993 | 0.92548338 |
| 10 | ANAC013 | 0.888188073 | 0.895478567 | 0.891789962 | 0.892538173 | 0.806984699 | 0.891789962 |
| 11 | ANAC016 | 0.938988717 | 0.932613391 | 0.935853866 | 0.937030859 | 0.879894684 | 0.935853866 |
| 12 | ANAC017 | 0.951419558 | 0.940813285 | 0.946148963 | 0.946740609 | 0.898082703 | 0.946148963 |
| 13 | ANAC020 | 0.917334458 | 0.930046355 | 0.923693086 | 0.923174873 | 0.859019613 | 0.923693086 |
| 14 | ANAC028 | 0.881777415 | 0.884615385 | 0.883193792 | 0.88321764 | 0.793674405 | 0.883193792 |
| 15 | ANAC034 | 0.896626768 | 0.861581921 | 0.87943459 | 0.883409274 | 0.787645499 | 0.87943459 |
| 16 | ANAC038 | 0.894792774 | 0.951754386 | 0.922827847 | 0.921729611 | 0.857413383 | 0.922827847 |
| 17 | ANAC042 | 0.909924488 | 0.953090508 | 0.931260229 | 0.930501931 | 0.871891528 | 0.931260229 |
| 18 | ANAC045 | 0.848627303 | 0.940412103 | 0.894815507 | 0.889085894 | 0.810876168 | 0.894815507 |
| 19 | ANAC046 | 0.941651376 | 0.955267482 | 0.948476251 | 0.948000369 | 0.902249353 | 0.948476251 |
| 20 | ANAC047 | 0.839185089 | 0.9805026 | 0.909859155 | 0.902985075 | 0.83435308 | 0.909859155 |
| 21 | ANAC050 | 0.916123908 | 0.935122336 | 0.925653207 | 0.92471739 | 0.86232824 | 0.925653207 |
| 22 | ANAC053 | 0.922325103 | 0.942400406 | 0.932430706 | 0.931307622 | 0.873947412 | 0.932430706 |
| 23 | ANAC055 | 0.886975717 | 0.981275078 | 0.933895297 | 0.930954588 | 0.876033214 | 0.933895297 |
| 24 | ANAC057 | 0.910088645 | 0.932639115 | 0.922822492 | 0.911242604 | 0.855091 | 0.922822492 |
| 25 | ANAC058 | 0.886344502 | 0.921344538 | 0.903860712 | 0.902064947 | 0.826095929 | 0.903860712 |
| 26 | ANAC062 | 0.914168937 | 0.971493359 | 0.947627151 | 0.935626307 | 0.897151793 | 0.947627151 |
| 27 | ANAC070 | 0.887152064 | 0.899731183 | 0.893504378 | 0.891860225 | 0.809642198 | 0.893504378 |
| 28 | ANAC071 | 0.870556578 | 0.939496271 | 0.90513834 | 0.901451826 | 0.827835018 | 0.90513834 |
| 29 | ANAC075 | 0.836349925 | 0.858895706 | 0.847528517 | 0.846888125 | 0.741506925 | 0.847528517 |
| 30 | ANAC079 | 0.921232877 | 0.969325153 | 0.944411591 | 0.944964871 | 0.894921684 | 0.944411591 |
| 31 | ANAC083 | 0.930204206 | 0.955112599 | 0.942667885 | 0.941902631 | 0.891874347 | 0.942667885 |
| 32 | ANAC087 | 0.925564682 | 0.946060286 | 0.935660328 | 0.935894108 | 0.879583137 | 0.935660328 |
| 33 | ANAC092 | 0.911311054 | 0.958319871 | 0.934751087 | 0.933355274 | 0.877895401 | 0.934751087 |
| 34 | ANAC094 | 0.867521368 | 0.928229665 | 0.896162528 | 0.898230088 | 0.813570237 | 0.896162528 |
| 35 | ANAC096 | 0.85612608 | 0.938791125 | 0.897390197 | 0.893131795 | 0.815230782 | 0.897390197 |
| 36 | ANAC103 | 0.926233512 | 0.955384615 | 0.942543577 | 0.934220251 | 0.889922621 | 0.942543577 |
| 37 | ANL2 | 0.994094488 | 0.98120915 | 0.98777065 | 0.988064958 | 0.975824825 | 0.98777065 |
| 38 | AREB3 | 0.958718861 | 0.989751098 | 0.974016601 | 0.973969631 | 0.949370546 | 0.974016601 |
| 39 | ARF2 | 0.744833525 | 0.884012539 | 0.814671815 | 0.800185014 | 0.695019765 | 0.814671815 |
| 40 | ASL18 | 0.908949819 | 0.962962963 | 0.935458377 | 0.934823091 | 0.87914062 | 0.935458377 |
| 41 | AT1G01250 | 0.964285714 | 0.933649289 | 0.949425287 | 0.95154185 | 0.903764931 | 0.949425287 |
| 42 | AT1G12630 | 0.994328577 | 0.986989188 | 0.990661906 | 0.990703609 | 0.981497592 | 0.990661906 |
| 43 | At1g19000 | 0.960147767 | 0.977831966 | 0.969033695 | 0.968605308 | 0.939969532 | 0.969033695 |
| 44 | AT1G19040 | 0.917218543 | 0.891525424 | 0.904522613 | 0.906710311 | 0.827158401 | 0.904522613 |
| 45 | At1g19210 | 0.988520971 | 0.99071161 | 0.989606533 | 0.989686165 | 0.979427859 | 0.989606533 |
| 46 | AT1G20910 | 0.985088537 | 0.98182388 | 0.983463339 | 0.983560794 | 0.967472377 | 0.983463339 |
| 47 | At1g22810 | 0.994947532 | 0.99276818 | 0.993875938 | 0.993981751 | 0.98782311 | 0.993875938 |
| 48 | AT1G24250 | 0.864022663 | 0.892744479 | 0.87761194 | 0.88150289 | 0.784856392 | 0.87761194 |
| 49 | At1g25550 | 0.973793405 | 0.986067416 | 0.979842729 | 0.98 | 0.960494021 | 0.979842729 |
| 50 | AT1G28160 | 0.99132948 | 0.994129159 | 0.992718447 | 0.99276411 | 0.985542329 | 0.992718447 |
| 51 | At1g36060 | 0.993447993 | 0.989166667 | 0.991325898 | 0.991418063 | 0.982800221 | 0.991325898 |
| 52 | AT1G44830 | 0.984126984 | 0.985324948 | 0.98470948 | 0.98510427 | 0.969865363 | 0.98470948 |
| 53 | AT1G47655 | 0.965286491 | 0.977243995 | 0.971242653 | 0.971176099 | 0.944136909 | 0.971242653 |
| 54 | At1g49010 | 0.970512821 | 0.990174289 | 0.980325763 | 0.98016597 | 0.96141943 | 0.980325763 |
| 55 | AT1G49560 | 0.946503791 | 0.976714309 | 0.963644647 | 0.957494407 | 0.928431217 | 0.963644647 |
| 56 | At1g64620 | 0.984963145 | 0.983471883 | 0.984215686 | 0.984189335 | 0.968929556 | 0.984215686 |
| 57 | At1g68670 | 0.970376762 | 0.991499008 | 0.981016272 | 0.980671414 | 0.962737467 | 0.981016272 |
| 58 | AT1G69570 | 0.980004818 | 0.965408039 | 0.972747093 | 0.973089343 | 0.946968357 | 0.972747093 |
| 59 | At1g69690 | 0.923076923 | 0.969798658 | 0.94639866 | 0.945205479 | 0.898439988 | 0.94639866 |
| 60 | AT1G71450 | 0.994187398 | 0.992030005 | 0.993113108 | 0.993148299 | 0.9863207 | 0.993113108 |
| 61 | At1g72010 | 0.960569551 | 0.989010989 | 0.97761194 | 0.971745152 | 0.954257631 | 0.97761194 |
| 62 | AT1G72740 | 0.982107049 | 0.985165522 | 0.983626911 | 0.98370027 | 0.96778924 | 0.983626911 |
| 63 | At1g74840 | 0.95077125 | 0.977086743 | 0.963946247 | 0.96341869 | 0.930465894 | 0.963946247 |
| 64 | AT1G76880 | 0.666666667 | 0.986486486 | 0.827118644 | 0.793522267 | 0.699417637 | 0.827118644 |
| 65 | AT1G77200 | 0.988507485 | 0.990903503 | 0.989829129 | 0.988656987 | 0.979647825 | 0.989829129 |
| 66 | At1g78700 | 0.978900256 | 0.98557377 | 0.982194885 | 0.982354828 | 0.965020499 | 0.982194885 |
| 67 | At2g01060 | 0.964502569 | 0.955690299 | 0.960093349 | 0.960241804 | 0.923369205 | 0.960093349 |
| 68 | At2g03500 | 0.977125032 | 0.984440928 | 0.980756644 | 0.980821918 | 0.96225292 | 0.980756644 |
| 69 | AT2G15740 | 0.989949749 | 0.983333333 | 0.986700767 | 0.986973948 | 0.973743486 | 0.986700767 |
| 70 | AT2G20110 | 0.979148279 | 0.982991714 | 0.981081377 | 0.98093396 | 0.962876243 | 0.981081377 |
| 71 | AT2G20400 | 0.966346154 | 0.975501114 | 0.97082879 | 0.971275168 | 0.943344309 | 0.97082879 |
| 72 | AT2G28810 | 0.973135525 | 0.967711429 | 0.970438054 | 0.970670577 | 0.942619946 | 0.970438054 |
| 73 | AT2G33550 | 0.941586074 | 0.951996928 | 0.946810561 | 0.946345257 | 0.899269253 | 0.946810561 |
| 74 | At2g33710 | 0.986650053 | 0.989762931 | 0.988199535 | 0.988233197 | 0.976677321 | 0.988199535 |
| 75 | AT2G40260 | 0.968664436 | 0.965307365 | 0.966986155 | 0.967046317 | 0.936151747 | 0.966986155 |
| 76 | At2g44940 | 0.995544239 | 0.996725606 | 0.996126533 | 0.996178344 | 0.992281652 | 0.996126533 |
| 77 | At2g45680 | 0.773049645 | 0.968992248 | 0.866666667 | 0.858267717 | 0.765603629 | 0.866666667 |
| 78 | At3g04030 | 0.962452885 | 0.977878699 | 0.970125328 | 0.970046756 | 0.94203166 | 0.970125328 |
| 79 | At3g09600 | 0.969378531 | 0.979239535 | 0.974312652 | 0.974166809 | 0.949942182 | 0.974312652 |
| 80 | AT3G09735 | 0.929481733 | 0.983739837 | 0.956512471 | 0.955458515 | 0.916699376 | 0.956512471 |
| 81 | AT3G10030 | 0.983710407 | 0.98336414 | 0.983539095 | 0.983710407 | 0.96761653 | 0.983539095 |
| 82 | AT3G10113 | 0.962786143 | 0.972008312 | 0.967400612 | 0.967226219 | 0.936923551 | 0.967400612 |
| 83 | AT3G10580 | 0.852531646 | 0.928664921 | 0.88996139 | 0.887351779 | 0.803723385 | 0.88996139 |
| 84 | At3g11280 | 0.972886762 | 0.964527027 | 0.968826907 | 0.969793323 | 0.939534817 | 0.968826907 |
| 85 | AT3G12130 | 0.992368493 | 0.987044534 | 0.989693518 | 0.989671106 | 0.979599237 | 0.989693518 |
| 86 | At3g12730 | 0.975354282 | 0.981273408 | 0.978294574 | 0.978368356 | 0.957530496 | 0.978294574 |
| 87 | At3g14180 | 0.994540943 | 0.993353783 | 0.993956182 | 0.994047619 | 0.987982581 | 0.993956182 |
| 88 | AT3G16280 | 0.992922144 | 0.98859127 | 0.990736104 | 0.990668348 | 0.981642787 | 0.990736104 |
| 89 | At3g24120 | 0.978031803 | 0.979948586 | 0.978987155 | 0.979032155 | 0.958857157 | 0.978987155 |
| 90 | AT3G25990 | 0.957524272 | 0.951653944 | 0.954658385 | 0.955784373 | 0.913372019 | 0.954658385 |
| 91 | AT3G42860 | 0.970666667 | 0.99713877 | 0.983436853 | 0.983783784 | 0.96740142 | 0.983436853 |
| 92 | At3g45610 | 0.976516634 | 0.988332058 | 0.983147274 | 0.9807149 | 0.966326868 | 0.983147274 |
| 93 | AT3G52440 | 0.970127866 | 0.981598432 | 0.975898335 | 0.97561246 | 0.952950525 | 0.975898335 |
| 94 | AT3G60490 | 0.962319911 | 0.985889343 | 0.974074074 | 0.973831776 | 0.949481051 | 0.974074074 |
| 95 | At3g60580 | 0.980351295 | 0.989329268 | 0.984786865 | 0.984896067 | 0.970034392 | 0.984786865 |
| 96 | AT4G00250 | 0.95924225 | 0.970361926 | 0.964821965 | 0.964502165 | 0.932111408 | 0.964821965 |
| 97 | At4g01280 | 0.960730843 | 0.97798913 | 0.969375255 | 0.969055151 | 0.940615554 | 0.969375255 |
| 98 | At4g16750 | 0.993677555 | 0.995066495 | 0.994365898 | 0.994411051 | 0.988794544 | 0.994365898 |
| 99 | AT4G18450 | 0.972222222 | 0.98089172 | 0.976261128 | 0.977653631 | 0.953467255 | 0.976261128 |
| 100 | At4g18890 | 0.946153846 | 0.969111969 | 0.95761079 | 0.957198444 | 0.918796904 | 0.95761079 |
| 101 | AT4G26030 | 0.954267405 | 0.959177592 | 0.956694653 | 0.957055215 | 0.917133453 | 0.956694653 |
| 102 | At4g31060 | 0.988328912 | 0.995527728 | 0.992234894 | 0.991484832 | 0.984469883 | 0.992234894 |
| 103 | At4g32800 | 0.989455185 | 0.994690265 | 0.992063492 | 0.992070485 | 0.984252838 | 0.992063492 |
| 104 | At4g36780 | 0.980021598 | 0.988713318 | 0.984271523 | 0.984540277 | 0.969027788 | 0.984271523 |
| 105 | AT4G37180 | 0.971224247 | 0.964050633 | 0.967621631 | 0.967599661 | 0.937339079 | 0.967621631 |
| 106 | At4g38000 | 0.989550974 | 0.969407802 | 0.979448198 | 0.979591837 | 0.959735029 | 0.979448198 |
| 107 | AT5G02460 | 0.966261808 | 0.969078996 | 0.967723453 | 0.966453611 | 0.937440815 | 0.967723453 |
| 108 | At5g04390 | 0.990951864 | 0.991007194 | 0.990979614 | 0.990951864 | 0.982121794 | 0.990979614 |
| 109 | AT5G05550 | 0.99225761 | 0.989625462 | 0.990941073 | 0.990949829 | 0.982046226 | 0.990941073 |
| 110 | At5g05790 | 0.963833635 | 0.97256386 | 0.968099861 | 0.968650613 | 0.938214308 | 0.968099861 |
| 111 | At5g08330 | 0.969162996 | 0.985731272 | 0.977129788 | 0.977777778 | 0.955264024 | 0.977129788 |
| 112 | At5g08520 | 0.978455071 | 0.982804233 | 0.980622199 | 0.980645161 | 0.961995152 | 0.980622199 |
| 113 | At5g08750 | 0.951557093 | 0.965753425 | 0.95869191 | 0.958188153 | 0.920781296 | 0.95869191 |
| 114 | At5g18450 | 0.97421981 | 0.990084986 | 0.981981982 | 0.982216142 | 0.964604701 | 0.981981982 |
| 115 | AT5G22990 | 0.964047936 | 0.983298539 | 0.973460361 | 0.973772697 | 0.948316739 | 0.973460361 |
| 116 | AT5G23930 | 0.996458087 | 0.992058644 | 0.994296007 | 0.994403535 | 0.98865285 | 0.994296007 |
| 117 | At5g29000 | 0.956645345 | 0.983050847 | 0.969890188 | 0.96939143 | 0.941568111 | 0.969890188 |
| 118 | AT5G45580 | 0.950959488 | 0.979464497 | 0.966992448 | 0.961848524 | 0.934978901 | 0.966992448 |
| 119 | At5g47390 | 0.979965655 | 0.987017946 | 0.983490791 | 0.983437051 | 0.967525944 | 0.983490791 |
| 120 | AT5G47660 | 0.955763726 | 0.94397463 | 0.949914776 | 0.950569358 | 0.904827046 | 0.949914776 |
| 121 | At5g52660 | 0.97308879 | 0.977743799 | 0.975426621 | 0.975261982 | 0.952058585 | 0.975426621 |
| 122 | AT5G56840 | 0.956394964 | 0.983205782 | 0.971273521 | 0.967357061 | 0.943379459 | 0.971273521 |
| 123 | At5g58900 | 0.96844703 | 0.97913013 | 0.973802198 | 0.973595605 | 0.948972505 | 0.973802198 |
| 124 | AT5G60130 | 0.923439135 | 0.94904014 | 0.936235171 | 0.935429732 | 0.880564072 | 0.936235171 |
| 125 | AT5G61620 | 0.964367102 | 0.981640559 | 0.973894463 | 0.9707017 | 0.948539832 | 0.973894463 |
| 126 | At5g62940 | 0.979285634 | 0.982329407 | 0.980813208 | 0.980713164 | 0.96236159 | 0.980813208 |
| 127 | AT5G63260 | 0.968995011 | 0.957446809 | 0.963230701 | 0.963501063 | 0.929159169 | 0.963230701 |
| 128 | At5g65130 | 0.957654723 | 0.986254296 | 0.971571906 | 0.971900826 | 0.944740096 | 0.971571906 |
| 129 | At5g66730 | 0.926023778 | 0.982197134 | 0.954306952 | 0.952661382 | 0.912618363 | 0.954306952 |
| 130 | AT5G66940 | 0.989149342 | 0.988232369 | 0.988690969 | 0.988698928 | 0.977637711 | 0.988690969 |
| 131 | ATAF1 | 0.866690883 | 0.955892986 | 0.911396992 | 0.907051891 | 0.837828232 | 0.911396992 |
| 132 | AtGRF6 | 0.976429288 | 0.984407484 | 0.980347116 | 0.980609418 | 0.961458932 | 0.980347116 |
| 133 | ATHB13 | 0.986626832 | 0.989726523 | 0.988163558 | 0.988243677 | 0.976606169 | 0.988163558 |
| 134 | ATHB15 | 0.976358149 | 0.990180879 | 0.983176141 | 0.983282675 | 0.966915341 | 0.983176141 |
| 135 | ATHB18 | 0.972256473 | 0.98310388 | 0.977639752 | 0.977681339 | 0.956277947 | 0.977639752 |
| 136 | ATHB20 | 0.98013245 | 0.991354467 | 0.985621652 | 0.985845129 | 0.971648734 | 0.985621652 |
| 137 | ATHB21 | 0.957252728 | 0.985868912 | 0.972800784 | 0.969828758 | 0.946557703 | 0.972800784 |
| 138 | ATHB23 | 0.970531886 | 0.980181503 | 0.976105137 | 0.971684326 | 0.95218674 | 0.976105137 |
| 139 | ATHB24 | 0.976160991 | 0.970137586 | 0.973164281 | 0.973373466 | 0.947765218 | 0.973164281 |
| 140 | ATHB25 | 0.96238076 | 0.967263702 | 0.964823952 | 0.964713805 | 0.932121555 | 0.964823952 |
| 141 | AtHB32 | 0.982688071 | 0.969408781 | 0.976072021 | 0.976311469 | 0.953282366 | 0.976072021 |
| 142 | ATHB33 | 0.945403235 | 0.957415991 | 0.951760437 | 0.948595077 | 0.907824626 | 0.951760437 |
| 143 | ATHB34 | 0.954764513 | 0.972537572 | 0.96370858 | 0.963151207 | 0.930030082 | 0.96370858 |
| 144 | ATHB40 | 0.988201982 | 0.985971037 | 0.98687795 | 0.983930082 | 0.973198922 | 0.98687795 |
| 145 | ATHB53 | 0.977099237 | 0.977684532 | 0.977391968 | 0.977379021 | 0.955806164 | 0.977391968 |
| 146 | ATHB5 | 0.988796323 | 0.982538086 | 0.985668103 | 0.985716843 | 0.971746409 | 0.985668103 |
| 147 | ATHB6 | 0.98481269 | 0.980835044 | 0.982837723 | 0.982988041 | 0.966261782 | 0.982837723 |
| 148 | ATHB7 | 0.96669779 | 0.988596769 | 0.97755102 | 0.977498033 | 0.956104259 | 0.97755102 |
| 149 | AtIDD11 | 0.933663366 | 0.984358707 | 0.958354495 | 0.958333333 | 0.920121954 | 0.958354495 |
| 150 | ATY13 | 0.939362795 | 0.965441301 | 0.952474236 | 0.951587715 | 0.90942081 | 0.952474236 |
| 151 | BAM8 | 0.98054755 | 0.976556777 | 0.978568834 | 0.97878461 | 0.958051781 | 0.978568834 |
| 152 | BBX31 | 0.973496835 | 0.985625873 | 0.979533035 | 0.979502488 | 0.959902253 | 0.979533035 |
| 153 | bHLH10 | 0.988204939 | 0.994592646 | 0.991434299 | 0.991310778 | 0.983011749 | 0.991434299 |
| 154 | bHLH122 | 0.971835653 | 0.993027888 | 0.982421227 | 0.982250502 | 0.965453413 | 0.982421227 |
| 155 | bHLH28 | 0.980898876 | 0.97493453 | 0.977915029 | 0.977968633 | 0.956804907 | 0.977915029 |
| 156 | bHLH31 | 0.992982456 | 0.990530303 | 0.991803279 | 0.992112182 | 0.983715944 | 0.991803279 |
| 157 | bHLH80 | 0.989631336 | 0.995195195 | 0.99235519 | 0.99248989 | 0.984822257 | 0.99235519 |
| 158 | BIM1 | 0.917525773 | 0.979487179 | 0.948586118 | 0.946808511 | 0.90225876 | 0.948586118 |
| 159 | BIM2 | 0.980564508 | 0.980154534 | 0.980340818 | 0.978414462 | 0.961145058 | 0.980340818 |
| 160 | BOS1 | 0.952023988 | 0.984326019 | 0.967816092 | 0.967987805 | 0.937684304 | 0.967816092 |
| 161 | BPC1 | 0.975632792 | 0.988544601 | 0.982107543 | 0.981939163 | 0.964850877 | 0.982107543 |
| 162 | bZIP16 | 0.957165733 | 0.981154771 | 0.969150641 | 0.968800648 | 0.940188515 | 0.969150641 |
| 163 | bZIP18 | 0.962614071 | 0.986068477 | 0.975628931 | 0.97234612 | 0.951764343 | 0.975628931 |
| 164 | bZIP28 | 0.944954128 | 0.977596741 | 0.96042471 | 0.961718021 | 0.923870042 | 0.96042471 |
| 165 | bZIP3 | 0.967588933 | 0.952595014 | 0.960216998 | 0.961130742 | 0.923553604 | 0.960216998 |
| 166 | bZIP44 | 0.952264382 | 0.98988622 | 0.970771144 | 0.970679975 | 0.943229299 | 0.970771144 |
| 167 | bZIP48 | 0.972435897 | 0.974051896 | 0.973228861 | 0.973684211 | 0.947875415 | 0.973228861 |
| 168 | bZIP50 | 0.976336678 | 0.976518709 | 0.976427173 | 0.976556063 | 0.953964308 | 0.976427173 |
| 169 | bZIP52 | 0.982189314 | 0.972154472 | 0.97721085 | 0.977494523 | 0.955452277 | 0.97721085 |
| 170 | bZIP53 | 0.973487032 | 0.973147972 | 0.973316756 | 0.973206569 | 0.94805662 | 0.973316756 |
| 171 | bZIP68 | 0.95879323 | 0.968253968 | 0.963515755 | 0.963401109 | 0.929691406 | 0.963515755 |
| 172 | BZR1 | 0.980645161 | 0.993355482 | 0.98690671 | 0.987012987 | 0.974153456 | 0.98690671 |
| 173 | CAMTA1 | 0.973593571 | 0.989937107 | 0.981392557 | 0.982049797 | 0.9634258 | 0.981392557 |
| 174 | CAMTA5 | 0.933920705 | 0.94092827 | 0.9375 | 0.93598234 | 0.882746061 | 0.9375 |
| 175 | CBF1 | 0.989409442 | 0.989558956 | 0.989484091 | 0.98949825 | 0.979189313 | 0.989484091 |
| 176 | CBF2 | 0.983293556 | 0.990543735 | 0.986935867 | 0.986826347 | 0.974210969 | 0.986935867 |
| 177 | CBF3 | 0.976315065 | 0.98254228 | 0.979443607 | 0.979281768 | 0.959729522 | 0.979443607 |
| 178 | CBF4 | 0.99358758 | 0.992272371 | 0.992928499 | 0.992917369 | 0.98595697 | 0.992928499 |
| 179 | CDF3 | 0.98361844 | 0.983213076 | 0.983417342 | 0.983547148 | 0.967382623 | 0.983417342 |
| 180 | CDM1 | 0.896742897 | 0.923455056 | 0.910010464 | 0.909346451 | 0.8361765 | 0.910010464 |
| 181 | CEJ1 | 0.99405558 | 0.994369618 | 0.994235764 | 0.993243745 | 0.988285442 | 0.994235764 |
| 182 | COG1 | 0.965992378 | 0.990005879 | 0.977983267 | 0.977744807 | 0.956924854 | 0.977983267 |
| 183 | CRC | 0.974716013 | 0.989920806 | 0.982386054 | 0.98209341 | 0.965381474 | 0.982386054 |
| 184 | CRF10 | 0.995863495 | 0.998430962 | 0.997139886 | 0.997152472 | 0.994296011 | 0.997139886 |
| 185 | CRF4 | 0.990920882 | 0.993045897 | 0.991946309 | 0.992207792 | 0.984004304 | 0.991946309 |
| 186 | CUC1 | 0.841286307 | 0.958980044 | 0.898177921 | 0.895143488 | 0.816232555 | 0.898177921 |
| 187 | CUC2 | 0.893324607 | 0.94548552 | 0.92125266 | 0.913348946 | 0.853635084 | 0.92125266 |
| 188 | CUC3 | 0.905906952 | 0.948650079 | 0.927143609 | 0.925995191 | 0.864806537 | 0.927143609 |
| 189 | DAG2 | 0.995696333 | 0.991195488 | 0.993194678 | 0.992365103 | 0.986320569 | 0.993194678 |
| 190 | DDF1 | 0.963838665 | 0.991643454 | 0.977731385 | 0.977433004 | 0.956438563 | 0.977731385 |
| 191 | DEAR2 | 0.994016242 | 0.993500867 | 0.99376031 | 0.99380386 | 0.987597874 | 0.99376031 |
| 192 | DEAR3 | 0.98496732 | 0.99606609 | 0.991124691 | 0.989981935 | 0.982175154 | 0.991124691 |
| 193 | DEAR5 | 0.967375887 | 0.986363636 | 0.976556777 | 0.977077364 | 0.954184376 | 0.976556777 |
| 194 | DEL2 | 0.961722488 | 0.995271868 | 0.978596908 | 0.97810219 | 0.958077258 | 0.978596908 |
| 195 | dof24 | 0.979194879 | 0.981262327 | 0.980227903 | 0.980220593 | 0.961237629 | 0.980227903 |
| 196 | dof42 | 0.93902439 | 0.970212766 | 0.954261954 | 0.954545455 | 0.912680314 | 0.954261954 |
| 197 | dof43 | 0.983526113 | 0.986924686 | 0.985229855 | 0.98516633 | 0.97089541 | 0.985229855 |
| 198 | dof45 | 0.981300701 | 0.971409029 | 0.976342316 | 0.976400575 | 0.953802566 | 0.976342316 |
| 199 | DREB19 | 0.975582374 | 0.99030837 | 0.983833148 | 0.981505012 | 0.967676818 | 0.983833148 |
| 200 | E2FA | 0.980561555 | 0.991469194 | 0.98637052 | 0.985349973 | 0.972981087 | 0.98637052 |
| 201 | EIN3 | 0.820580475 | 0.821325648 | 0.820936639 | 0.82712766 | 0.705603091 | 0.820936639 |
| 202 | EPR1 | 0.960885135 | 0.987995726 | 0.97697933 | 0.971364923 | 0.953222588 | 0.97697933 |
| 203 | ERF105 | 0.978756708 | 0.989545455 | 0.984107304 | 0.984148398 | 0.968718599 | 0.984107304 |
| 204 | ERF10 | 0.98079561 | 0.984604619 | 0.982680984 | 0.982817869 | 0.965959557 | 0.982680984 |
| 205 | ERF115 | 0.981313703 | 0.988296135 | 0.984873716 | 0.984519244 | 0.970189118 | 0.984873716 |
| 206 | ERF11 | 0.990401396 | 0.993079585 | 0.991746308 | 0.991699432 | 0.983628312 | 0.991746308 |
| 207 | ERF13 | 0.96882662 | 0.982285516 | 0.975584234 | 0.975317348 | 0.952353075 | 0.975584234 |
| 208 | ERF15 | 0.97598411 | 0.990201135 | 0.984278622 | 0.981032762 | 0.968118389 | 0.984278622 |
| 209 | ERF2 | 0.964125561 | 0.981220657 | 0.972477064 | 0.972850679 | 0.946454535 | 0.972477064 |
| 210 | ERF38 | 0.992788462 | 0.988463874 | 0.99063727 | 0.990704648 | 0.981448804 | 0.99063727 |
| 211 | ERF3 | 0.987390883 | 0.991683992 | 0.989463121 | 0.989790958 | 0.97912665 | 0.989463121 |
| 212 | ERF4 | 0.980032823 | 0.992327926 | 0.986858116 | 0.985152598 | 0.973714811 | 0.986858116 |
| 213 | ERF5 | 0.969111969 | 0.987951807 | 0.978346457 | 0.978557505 | 0.957622473 | 0.978346457 |
| 214 | ERF7 | 0.972065992 | 0.980689133 | 0.976356443 | 0.976367574 | 0.953830007 | 0.976356443 |
| 215 | ERF8 | 0.984238672 | 0.995072614 | 0.989877176 | 0.989390296 | 0.979916605 | 0.989877176 |
| 216 | ESE1 | 0.971922246 | 0.995505618 | 0.983480176 | 0.983606557 | 0.967499477 | 0.983480176 |
| 217 | ESE3 | 0.995322058 | 0.993612491 | 0.994461319 | 0.994427467 | 0.988983576 | 0.994461319 |
| 218 | FAR1 | 0.982517483 | 0.975778547 | 0.979130435 | 0.979094077 | 0.959131329 | 0.979130435 |
| 219 | FRS9 | 0.974634566 | 0.985370052 | 0.98 | 0.979900584 | 0.960797909 | 0.98 |
| 220 | FUS3 | 0.980276134 | 0.976744186 | 0.978571429 | 0.979310345 | 0.958007622 | 0.978571429 |
| 221 | GATA11 | 0.979035639 | 0.989316239 | 0.984126984 | 0.984193888 | 0.96875644 | 0.984126984 |
| 222 | GATA1 | 0.965517241 | 0.914383562 | 0.939862543 | 0.941176471 | 0.88682706 | 0.939862543 |
| 223 | GATA20 | 0.987590683 | 0.991467577 | 0.989535769 | 0.989479725 | 0.979289875 | 0.989535769 |
| 224 | GATA4 | 0.972166998 | 0.982758621 | 0.977249224 | 0.978 | 0.955480169 | 0.977249224 |
| 225 | GBF3 | 0.952309237 | 0.966548192 | 0.960077564 | 0.955908289 | 0.922650402 | 0.960077564 |
| 226 | GBF5 | 0.973643411 | 0.987798678 | 0.980779088 | 0.980483997 | 0.962286655 | 0.980779088 |
| 227 | GBF6 | 0.963198394 | 0.968792402 | 0.965976082 | 0.966107383 | 0.934265828 | 0.965976082 |
| 228 | GRF9 | 0.980474198 | 0.987288136 | 0.983859649 | 0.983904829 | 0.968239679 | 0.983859649 |
| 229 | GT1 | 0.950617284 | 0.926666667 | 0.939102564 | 0.941896024 | 0.885346894 | 0.939102564 |
| 230 | GT2 | 0.940370281 | 0.968322253 | 0.955785614 | 0.950194363 | 0.914396873 | 0.955785614 |
| 231 | GT3a | 0.726342711 | 0.865 | 0.796460177 | 0.77914952 | 0.672330582 | 0.796460177 |
| 232 | GTL1 | 0.958731169 | 0.93211903 | 0.945413111 | 0.94608459 | 0.896751299 | 0.945413111 |
| 233 | HAT2 | 0.966556038 | 0.987714988 | 0.977841315 | 0.976024749 | 0.956411744 | 0.977841315 |
| 234 | HDG1 | 0.981423264 | 0.988538682 | 0.984976391 | 0.984941919 | 0.970403667 | 0.984976391 |
| 235 | HSF21 | 0.949541284 | 0.957627119 | 0.953744493 | 0.951724138 | 0.911612913 | 0.953744493 |
| 236 | HSF3 | 0.971777269 | 0.958122411 | 0.96496864 | 0.965297772 | 0.932382534 | 0.96496864 |
| 237 | HSF6 | 0.945945946 | 0.972752044 | 0.959294437 | 0.95890411 | 0.921881361 | 0.959294437 |
| 238 | HSF7 | 0.969402985 | 0.980654762 | 0.975037258 | 0.974859287 | 0.951316835 | 0.975037258 |
| 239 | HSFA6B | 0.92770683 | 0.973299126 | 0.952276102 | 0.947165533 | 0.908221858 | 0.952276102 |
| 240 | HSFB3 | 0.877777778 | 0.970845481 | 0.923186344 | 0.921282799 | 0.857759826 | 0.923186344 |
| 241 | HSFC1 | 0.921000397 | 0.956820016 | 0.938763258 | 0.938131824 | 0.884975485 | 0.938763258 |
| 242 | HY5 | 0.975057434 | 0.989463286 | 0.982248521 | 0.98214876 | 0.965124361 | 0.982248521 |
| 243 | IDD4 | 0.900472689 | 0.977150538 | 0.938363443 | 0.936629336 | 0.884063395 | 0.938363443 |
| 244 | IDD5 | 0.880899705 | 0.962694676 | 0.922163347 | 0.918139892 | 0.855869418 | 0.922163347 |
| 245 | IDD7 | 0.941885188 | 0.985925405 | 0.963983051 | 0.963043478 | 0.930482868 | 0.963983051 |
| 246 | JKD | 0.897590361 | 0.982594937 | 0.93904321 | 0.937844217 | 0.885254499 | 0.93904321 |
| 247 | LBD13 | 0.942133816 | 0.948352627 | 0.945266936 | 0.944696283 | 0.896513521 | 0.945266936 |
| 248 | LBD18 | 0.920072661 | 0.946539849 | 0.933982591 | 0.929698972 | 0.876210858 | 0.933982591 |
| 249 | LBD19 | 0.920801794 | 0.917361111 | 0.919073531 | 0.918869772 | 0.85124366 | 0.919073531 |
| 250 | LBD23 | 0.924276169 | 0.954976303 | 0.939150402 | 0.93997735 | 0.885650632 | 0.939150402 |
| 251 | LBD2 | 0.95281457 | 0.948990726 | 0.950891632 | 0.950715859 | 0.906604891 | 0.950891632 |
| 252 | LCL1 | 0.956658786 | 0.989935588 | 0.97311828 | 0.972951312 | 0.947664847 | 0.97311828 |
| 253 | LEP | 0.929411765 | 0.943037975 | 0.93597561 | 0.93768546 | 0.88004923 | 0.93597561 |
| 254 | LHY1 | 0.945389188 | 0.973128409 | 0.961542903 | 0.953562962 | 0.923792227 | 0.961542903 |
| 255 | LMI1 | 0.994022895 | 0.991600032 | 0.992674816 | 0.991762268 | 0.985276307 | 0.992674816 |
| 256 | LOB | 0.986725664 | 0.981176471 | 0.984036488 | 0.984547461 | 0.9685481 | 0.984036488 |
| 257 | MGP | 0.965669014 | 0.986690328 | 0.97613787 | 0.975978648 | 0.953407265 | 0.97613787 |
| 258 | MP | 0.946584939 | 0.957885529 | 0.95223806 | 0.951943641 | 0.909032233 | 0.95223806 |
| 259 | MS188 | 0.960551885 | 0.97253825 | 0.966516986 | 0.966467885 | 0.935273609 | 0.966516986 |
| 260 | MYB101 | 0.948459896 | 0.965262834 | 0.956827924 | 0.95662873 | 0.917375692 | 0.956827924 |
| 261 | MYB107 | 0.932484454 | 0.953488372 | 0.943927753 | 0.938039917 | 0.893176452 | 0.943927753 |
| 262 | MYB116 | 0.967403522 | 0.987813885 | 0.977682723 | 0.977289932 | 0.956344162 | 0.977682723 |
| 263 | MYB118 | 0.964368706 | 0.992260062 | 0.978319783 | 0.978004713 | 0.957562755 | 0.978319783 |
| 264 | MYB119 | 0.965363366 | 0.981629566 | 0.973427622 | 0.973430469 | 0.948263813 | 0.973427622 |
| 265 | MYB121 | 0.979365079 | 0.988543372 | 0.983883965 | 0.984051037 | 0.968283165 | 0.983883965 |
| 266 | MYB13 | 0.950177936 | 0.981179423 | 0.965243902 | 0.965641953 | 0.932876643 | 0.965243902 |
| 267 | MYB27 | 0.962962963 | 0.993189103 | 0.980710421 | 0.976314269 | 0.960819035 | 0.980710421 |
| 268 | MYB33 | 0.964858109 | 0.968242135 | 0.966538348 | 0.966705952 | 0.935314181 | 0.966538348 |
| 269 | MYB3R1 | 0.926843834 | 0.95620219 | 0.941488964 | 0.940748598 | 0.889783673 | 0.941488964 |
| 270 | MYB3R4 | 0.973770492 | 0.949033392 | 0.961832061 | 0.96350365 | 0.926414867 | 0.961832061 |
| 271 | MYB3R5 | 0.954707986 | 0.98343949 | 0.968596059 | 0.969147005 | 0.939130841 | 0.968596059 |
| 272 | MYB44 | 0.947032546 | 0.949010989 | 0.948005621 | 0.948748002 | 0.901397053 | 0.948005621 |
| 273 | MYB49 | 0.935602269 | 0.956696878 | 0.946117805 | 0.945699831 | 0.898024465 | 0.946117805 |
| 274 | MYB55 | 0.971770176 | 0.981125828 | 0.976454983 | 0.976309643 | 0.954015981 | 0.976454983 |
| 275 | MYB56 | 0.96002161 | 0.974040632 | 0.966878278 | 0.967338051 | 0.935934153 | 0.966878278 |
| 276 | MYB57 | 0.962576851 | 0.988111321 | 0.975275464 | 0.975088004 | 0.951762792 | 0.975275464 |
| 277 | MYB58 | 0.959269663 | 0.976947902 | 0.968696222 | 0.96622458 | 0.939023464 | 0.968696222 |
| 278 | MYB61 | 0.980090158 | 0.985926065 | 0.983009481 | 0.98295187 | 0.966595653 | 0.983009481 |
| 279 | MYB62 | 0.965349076 | 0.984778122 | 0.9750386 | 0.974857439 | 0.95131617 | 0.9750386 |
| 280 | MYB63 | 0.966625464 | 0.986093552 | 0.97625 | 0.97627965 | 0.953623415 | 0.97625 |
| 281 | MYB65 | 0.956772334 | 0.973478939 | 0.965554948 | 0.963435868 | 0.933256037 | 0.965554948 |
| 282 | MYB67 | 0.969196732 | 0.986666667 | 0.977967969 | 0.977682403 | 0.956896505 | 0.977967969 |
| 283 | MYB70 | 0.959976798 | 0.900028645 | 0.929816977 | 0.93147601 | 0.869288731 | 0.929816977 |
| 284 | MYB73 | 0.939992814 | 0.930755712 | 0.935323383 | 0.934953538 | 0.879005548 | 0.935323383 |
| 285 | MYB74 | 0.96763754 | 0.989010989 | 0.978225367 | 0.978189749 | 0.957393841 | 0.978225367 |
| 286 | MYB77 | 0.92997919 | 0.959091464 | 0.944553124 | 0.943668095 | 0.895207189 | 0.944553124 |
| 287 | MYB81 | 0.940816327 | 0.933263816 | 0.93708097 | 0.937945066 | 0.882055553 | 0.93708097 |
| 288 | MYB83 | 0.975051499 | 0.982690976 | 0.978855436 | 0.978860294 | 0.958604422 | 0.978855436 |
| 289 | MYB88 | 0.993907746 | 0.991955307 | 0.992944549 | 0.993043478 | 0.985985811 | 0.992944549 |
| 290 | MYB93 | 0.933020478 | 0.937713311 | 0.935366894 | 0.935214881 | 0.879087334 | 0.935366894 |
| 291 | MYB96 | 0.876436782 | 0.917888563 | 0.896952104 | 0.895741557 | 0.815029445 | 0.896952104 |
| 292 | MYB98 | 0.977218225 | 0.978404319 | 0.977811094 | 0.977804439 | 0.956606864 | 0.977811094 |
| 293 | MYB99 | 0.949760766 | 0.981387479 | 0.965106732 | 0.965545197 | 0.932618631 | 0.965106732 |
| 294 | NAC2 | 0.920799093 | 0.932749364 | 0.926782683 | 0.926245279 | 0.864275034 | 0.926782683 |
| 295 | NAM | 0.916666667 | 0.974025974 | 0.949286066 | 0.939730837 | 0.901209319 | 0.949286066 |
| 296 | NAP | 0.913793103 | 0.961373391 | 0.937268396 | 0.936546185 | 0.882320886 | 0.937268396 |
| 297 | NGA4 | 0.941944848 | 0.969325153 | 0.955257271 | 0.955817378 | 0.914485666 | 0.955257271 |
| 298 | NLP7 | 0.827586207 | 0.869230769 | 0.848368522 | 0.845401174 | 0.742511858 | 0.848368522 |
| 299 | NST1 | 0.845301894 | 0.933126182 | 0.895307692 | 0.874272517 | 0.807015048 | 0.895307692 |
| 300 | NTL8 | 0.916666667 | 0.927419355 | 0.921927847 | 0.923034176 | 0.856009333 | 0.921927847 |
| 301 | NTM1 | 0.874681393 | 0.935850676 | 0.905112605 | 0.902575342 | 0.82795086 | 0.905112605 |
| 302 | NTM2 | 0.872070931 | 0.914452366 | 0.893017297 | 0.891839378 | 0.808806494 | 0.893017297 |
| 303 | OBP1 | 0.988986162 | 0.993135993 | 0.991047957 | 0.991085326 | 0.982255801 | 0.991047957 |
| 304 | OBP3 | 0.979616858 | 0.969565889 | 0.974611479 | 0.974836053 | 0.950506973 | 0.974611479 |
| 305 | OBP4 | 0.967199427 | 0.98489893 | 0.97606288 | 0.975812274 | 0.9532631 | 0.97606288 |
| 306 | PDF2 | 0.942771084 | 0.981012658 | 0.961419753 | 0.961597542 | 0.925784933 | 0.961419753 |
| 307 | PHV | 0.962987013 | 0.984794565 | 0.973910225 | 0.973576235 | 0.949167726 | 0.973910225 |
| 308 | PLT3 | 0.920731707 | 0.745341615 | 0.833846154 | 0.848314607 | 0.718352146 | 0.833846154 |
| 309 | PUCHI | 0.968215159 | 0.992471769 | 0.980185759 | 0.98019802 | 0.961150753 | 0.980185759 |
| 310 | Rap210 | 0.988210637 | 0.987977558 | 0.988095238 | 0.988210637 | 0.976471669 | 0.988095238 |
| 311 | RAP211 | 0.991127462 | 0.991104675 | 0.991115812 | 0.990913025 | 0.982380706 | 0.991115812 |
| 312 | RAP212 | 0.964161274 | 0.995346432 | 0.979457266 | 0.979519595 | 0.959747896 | 0.979457266 |
| 313 | RAP21 | 0.983154671 | 0.990147783 | 0.986529319 | 0.986933128 | 0.97339576 | 0.986529319 |
| 314 | RAP26 | 0.984013462 | 0.989075302 | 0.986639676 | 0.98608769 | 0.973594686 | 0.986639676 |
| 315 | RAV1 | 0.927572202 | 0.96435059 | 0.945903124 | 0.945057471 | 0.897599803 | 0.945903124 |
| 316 | REM19 | 0.982360699 | 0.989642887 | 0.985992314 | 0.985977249 | 0.972376654 | 0.985992314 |
| 317 | RKD2 | 0.826619401 | 0.911735861 | 0.868873742 | 0.863927248 | 0.771405059 | 0.868873742 |
| 318 | RRTF1 | 0.97871179 | 0.992080033 | 0.986291657 | 0.984083425 | 0.97242869 | 0.986291657 |
| 319 | RVE1 | 0.967182853 | 0.986777547 | 0.978003306 | 0.975233959 | 0.956428455 | 0.978003306 |
| 320 | SGR5 | 0.948911517 | 0.985397607 | 0.967135325 | 0.966559371 | 0.936390891 | 0.967135325 |
| 321 | SMB | 0.832298801 | 0.951091921 | 0.899191097 | 0.87826087 | 0.812888783 | 0.899191097 |
| 322 | SND2 | 0.877117467 | 0.863566755 | 0.87035106 | 0.871370439 | 0.774295801 | 0.87035106 |
| 323 | SND3 | 0.891140537 | 0.89798461 | 0.894559567 | 0.89428089 | 0.811350862 | 0.894559567 |
| 324 | SOL1 | 0.983272506 | 0.980232914 | 0.98175851 | 0.981854073 | 0.964181456 | 0.98175851 |
| 325 | SPL13 | 0.901639344 | 0.970760234 | 0.935028249 | 0.934844193 | 0.878334125 | 0.935028249 |
| 326 | SPL14 | 0.97265625 | 0.973333333 | 0.972990777 | 0.973289902 | 0.947433936 | 0.972990777 |
| 327 | SPL15 | 0.983493347 | 0.970588235 | 0.976978897 | 0.976911494 | 0.955015351 | 0.976978897 |
| 328 | SPL1 | 0.988946015 | 0.966163457 | 0.977625453 | 0.978009406 | 0.956233337 | 0.977625453 |
| 329 | SPL5 | 0.986096731 | 0.975678664 | 0.980405315 | 0.97857038 | 0.96129355 | 0.980405315 |
| 330 | SPL9 | 0.97541972 | 0.97824 | 0.97693398 | 0.975103157 | 0.954687006 | 0.97693398 |
| 331 | SRS7 | 0.973637961 | 0.977245863 | 0.975433951 | 0.975495231 | 0.952074409 | 0.975433951 |
| 332 | STOP1 | 0.879807692 | 0.912060302 | 0.895577396 | 0.895960832 | 0.812896883 | 0.895577396 |
| 333 | STZ | 0.98541552 | 0.967205585 | 0.976317239 | 0.976547587 | 0.953747958 | 0.976317239 |
| 334 | SVP | 0.868229588 | 0.896902227 | 0.882580958 | 0.88076103 | 0.792646261 | 0.882580958 |
| 335 | TBP3 | 0.981103166 | 0.9958159 | 0.988372093 | 0.988422948 | 0.97701287 | 0.988372093 |
| 336 | TCP20 | 0.938028169 | 0.942196532 | 0.940085592 | 0.940677966 | 0.887338507 | 0.940085592 |
| 337 | TCP24 | 0.966052376 | 0.990825688 | 0.978131213 | 0.978388998 | 0.957205723 | 0.978131213 |
| 338 | TCP3 | 0.969267139 | 0.978922717 | 0.974117647 | 0.973871734 | 0.949569506 | 0.974117647 |
| 339 | TCP7 | 0.934210526 | 0.989932886 | 0.96179402 | 0.96108291 | 0.926424689 | 0.96179402 |
| 340 | TCX2 | 0.974719545 | 0.980150582 | 0.977629456 | 0.975875979 | 0.956027435 | 0.977629456 |
| 341 | TGA10 | 0.962184874 | 0.981028152 | 0.971515152 | 0.971515152 | 0.944647871 | 0.971515152 |
| 342 | TGA1 | 0.977334053 | 0.981710597 | 0.979525862 | 0.97944835 | 0.959889348 | 0.979525862 |
| 343 | TGA2 | 0.980211873 | 0.971099434 | 0.975680836 | 0.975920398 | 0.952538894 | 0.975680836 |
| 344 | TGA3 | 0.943449576 | 0.984158416 | 0.963302752 | 0.963426372 | 0.929266154 | 0.963302752 |
| 345 | TGA4 | 0.947178478 | 0.977895084 | 0.962493831 | 0.962012662 | 0.927772301 | 0.962493831 |
| 346 | TGA5 | 0.942675159 | 0.975578406 | 0.959053103 | 0.958549223 | 0.921426225 | 0.959053103 |
| 347 | TGA6 | 0.959875021 | 0.973758531 | 0.967967625 | 0.961535294 | 0.936203312 | 0.967967625 |
| 348 | TGA9 | 0.978966132 | 0.972098015 | 0.975537898 | 0.975661752 | 0.952270801 | 0.975537898 |
| 349 | TINY | 0.969964664 | 0.997995992 | 0.983098592 | 0.983870968 | 0.966685586 | 0.983098592 |
| 350 | TRP1 | 0.968553459 | 0.983766234 | 0.976038339 | 0.976228209 | 0.953219039 | 0.976038339 |
| 351 | TRP2 | 0.978586724 | 0.967342342 | 0.973106476 | 0.973894513 | 0.947610501 | 0.973106476 |
| 352 | VIP1 | 0.958592133 | 0.988399072 | 0.972647702 | 0.973711882 | 0.94669082 | 0.972647702 |
| 353 | VND1 | 0.880785414 | 0.867285383 | 0.874010247 | 0.874448828 | 0.779753164 | 0.874010247 |
| 354 | VND2 | 0.8334 | 0.940698619 | 0.886638452 | 0.881065652 | 0.797944972 | 0.886638452 |
| 355 | VND3 | 0.860509554 | 0.934816348 | 0.901512989 | 0.886773876 | 0.81925511 | 0.901512989 |
| 356 | VND4 | 0.870481928 | 0.889341229 | 0.880620772 | 0.870856651 | 0.788535497 | 0.880620772 |
| 357 | VND6 | 0.856994819 | 0.88787234 | 0.872230971 | 0.871719195 | 0.777042957 | 0.872230971 |
| 358 | VRN1 | 0.996680891 | 0.993358634 | 0.995020157 | 0.995029586 | 0.990089848 | 0.995020157 |
| 359 | WIP5 | 0.736723672 | 0.9268 | 0.837357052 | 0.809995052 | 0.720006464 | 0.837357052 |
| 360 | WRKY14 | 0.983606557 | 0.990039841 | 0.986819199 | 0.986792923 | 0.973985491 | 0.986819199 |
| 361 | WRKY15 | 0.989553764 | 0.993204983 | 0.991381754 | 0.991354795 | 0.982911834 | 0.991381754 |
| 362 | WRKY18 | 0.98233922 | 0.976564203 | 0.979424548 | 0.979293544 | 0.959693798 | 0.979424548 |
| 363 | WRKY20 | 0.976154993 | 0.97412481 | 0.975150602 | 0.975428146 | 0.951529979 | 0.975150602 |
| 364 | WRKY21 | 0.974895397 | 0.997752809 | 0.985915493 | 0.986243386 | 0.972208153 | 0.985915493 |
| 365 | WRKY22 | 0.98962766 | 0.991560807 | 0.990593247 | 0.990593664 | 0.98136345 | 0.990593247 |
| 366 | WRKY24 | 0.980843841 | 0.996987418 | 0.990170984 | 0.988272661 | 0.980012794 | 0.990170984 |
| 367 | WRKY25 | 0.988955506 | 0.99408 | 0.991499841 | 0.991536819 | 0.98314375 | 0.991499841 |
| 368 | WRKY27 | 0.985552408 | 0.991354467 | 0.988428571 | 0.988492684 | 0.977124023 | 0.988428571 |
| 369 | WRKY28 | 0.970355731 | 0.98198865 | 0.976175781 | 0.976021866 | 0.953483269 | 0.976175781 |
| 370 | WRKY29 | 0.988310308 | 0.988485385 | 0.988397839 | 0.988397839 | 0.977064898 | 0.988397839 |
| 371 | WRKY30 | 0.988653555 | 0.996930161 | 0.992761905 | 0.992783897 | 0.985628204 | 0.992761905 |
| 372 | WRKY31 | 0.982309125 | 0.989215686 | 0.985673352 | 0.985981308 | 0.971743176 | 0.985673352 |
| 373 | WRKY33 | 0.984642646 | 0.988574865 | 0.98659118 | 0.98668245 | 0.973540595 | 0.98659118 |
| 374 | WRKY3 | 0.965968586 | 0.976982097 | 0.971539457 | 0.971052632 | 0.944681806 | 0.971539457 |
| 375 | WRKY40 | 0.991584853 | 0.991079812 | 0.991332865 | 0.99135312 | 0.982815873 | 0.991332865 |
| 376 | WRKY45 | 0.985458952 | 0.994232698 | 0.990259295 | 0.989204805 | 0.980522498 | 0.990259295 |
| 377 | WRKY50 | 0.976115038 | 0.975460123 | 0.975788701 | 0.975877193 | 0.952749138 | 0.975788701 |
| 378 | WRKY55 | 0.988644689 | 0.989966555 | 0.989300867 | 0.989369501 | 0.978829783 | 0.989300867 |
| 379 | WRKY65 | 0.980130902 | 0.994178525 | 0.988434334 | 0.985776419 | 0.976309991 | 0.988434334 |
| 380 | WRKY6 | 0.982619491 | 0.998104864 | 0.990294302 | 0.990303409 | 0.98077581 | 0.990294302 |
| 381 | WRKY70 | 0.982293763 | 0.986596263 | 0.984435011 | 0.984472676 | 0.969354228 | 0.984435011 |
| 382 | WRKY71 | 0.975746924 | 0.977043478 | 0.976398601 | 0.976261649 | 0.953909708 | 0.976398601 |
| 383 | WRKY75 | 0.996055894 | 0.992193599 | 0.99411468 | 0.994095484 | 0.988298465 | 0.99411468 |
| 384 | WRKY7 | 0.978405316 | 1 | 0.988754325 | 0.989084803 | 0.977738586 | 0.988754325 |
| 385 | WRKY8 | 0.96835443 | 0.985700127 | 0.976998518 | 0.976872872 | 0.95505041 | 0.976998518 |
Sheet 4: Ten fold Independent Random Trials (Dataset A)
| Fold | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|
| 1 | 95.3315 | 95.8015 | 95.5659 | 95.6131 |
| 2 | 92.1596 | 92.6429 | 92.3996 | 92.4491 |
| 3 | 92.0298 | 93.0196 | 92.5237 | 92.6681 |
| 4 | 92.5889 | 93.0903 | 92.8397 | 92.7636 |
| 5 | 92.271 | 93.062 | 92.6652 | 92.7084 |
| 6 | 92.1012 | 93.0565 | 92.5769 | 92.6003 |
| 7 | 94.2799 | 94.7739 | 94.5261 | 94.4525 |
| 8 | 94.2165 | 94.771 | 94.4932 | 94.3799 |
| 9 | 94.1726 | 94.9692 | 94.5697 | 94.5423 |
| 10 | 94.2107 | 94.9194 | 94.5643 | 94.6109 |
Sheet 5: Ten fold Independent Random Trials (Dataset B)
| Fold | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|
| 1 | 95.5294 | 96.8277 | 96.1785 | 98.6875 |
| 2 | 94.9736 | 96.3576 | 95.6651 | 95.6595 |
| 3 | 95.0142 | 96.2554 | 95.635 | 95.6446 |
| 4 | 95.0423 | 96.4525 | 95.7477 | 95.7448 |
| 5 | 95.0633 | 96.3278 | 95.6951 | 95.6978 |
| 6 | 95.0493 | 96.1965 | 95.6229 | 95.6209 |
| 7 | 95.0097 | 96.331 | 95.6702 | 95.6681 |
| 8 | 95.0623 | 96.34 | 95.7012 | 95.7058 |
| 9 | 94.9817 | 96.2908 | 95.6362 | 95.6361 |
| 10 | 95.0474 | 96.3175 | 95.6827 | 95.6834 |
Sheet 6: Average benchmarking details of tool on Dataset B
| S.No. | Tool | Sensitivity | Specificity | Accuracy | F1-Score | AUC |
|---|---|---|---|---|---|---|
| 1 | SeqConv | 83.74 | 88.86 | 86.3 | 85.72 | 76.78 |
| 2 | TSPTFBS | 97.9 | 63.02 | 80.46 | 83.92 | 67.15 |
| 3 | k-mer grammar | 86 | 85.17 | 85.59 | 85.73 | 76.8 |
| 4 | Wimtrap | 80.18 | 78.9 | 79.54 | 79.67 | 68.01 |
| 5 | PTFSpot | 95.52 | 96.44 | 95.98 | 95.96 | 92.31 |
| 6 | DNABERT | 78.17 | 80.92 | 79.55 | 79.27 | 68.02 |
| 7 | PlantBind | 61.49 | 65.05 | 63.27 | 60.35 | 51.44 |
| 8 | TSPTFBS 2.0 | 95.11 | 96.92 | 96.02 | 95.79 | 91.85 |
Sheet 7: Average benchmarking details of tool on Dataset C
| S.No. | Tool | Sensitivity | Specificity | Accuracy | F1-Score | AUC |
|---|---|---|---|---|---|---|
| 1 | SeqConv | 92.1433627 | 89.6492686 | 90.9072841 | 91.0038715 | 83.8610506 |
| 2 | TSPTFBS | 88.8752293 | 92.8119698 | 90.8388352 | 90.5133395 | 84.1822036 |
| 3 | k-mer grammar | 88.5026152 | 89.2773157 | 88.9285481 | 88.7489799 | 80.6523136 |
| 4 | Wimtrap | 82.2310219 | 81.3028963 | 81.6702776 | 80.7014562 | 70.5099586 |
| 5 | PTFSpot | 95.16611 | 96.8914336 | 96.039959 | 95.9406773 | 92.6172198 |
| 6 | DNABERT | 88.90220987 | 92.95298663 | 90.92864047 | 89.64411058 | 82.09069351 |
| 7 | PlantBind | 65.63 | 69.59 | 67.61 | 68.33 | 59.42 |
| 8 | TSPTFBS 2.0 | 95.50730376 | 96.30390081 | 95.91223488 | 95.79941971 | 92.21699399 |
Sheet 8: Sequence-based comparative details for the common TFs studied between A. thaliana and Z. mays.
| S.No. | TF Name | Number of amino acids in Arabidopsis thaliana | Number of amino acids in Zea mays | Structural comparision RMSD (Å) | Sequence identity between A. thaliana and Z. mays TFs in % | Number of overlaped residues |
|---|---|---|---|---|---|---|
| 1 | LHY1 | 645 | 663 | 1.03 | 39 | 387 |
| 2 | MYB56 | 323 | 242 | 0.82 | 40.7 | 108 |
| 3 | MYB62 | 286 | 306 | 0.72 | 77.2 | 145 |
| 4 | MYB81 | 427 | 304 | 1.29 | 35.7 | 28 |
| 5 | MYB88 | 484 | 228 | 0.91 | 36.2 | 105 |
| 6 | WRKY25 | 393 | 333 | 0.62 | 43.9 | 57 |
Sheet 9: Domain-based comparative details for the common TFs studied between A. thaliana and Z. mays.
| S.No. | TF Name | Domain Name | Domain Number (Arabidopsis thaliana) | Domain Number (Zea mays) | Domain Sequence (Arabidopsis thaliana) | Domain Sequence (Zea mays) | Number of amino acids in Arabidopsis thaliana | Number of amino acids in Zea mays | Domain sequence identity between At and Zm (Domain 1) | Domain sequence identity between At and Zm (Domain 2) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LHY1 | HTH DNA-binding domain | 1 | 0 | TITKQRERWTEDEHERFLEALRLYGRAWQRIEEHIGTKTAVQIRSHAQKFFTKLE | NA | 55 | NA | NA | NA |
| 2 | MYB56 | HTH DNA-binding domain | 2 | 2 | TKVCSRGHWRPTEDAKLKELVAQFGPQNWNLISNHLLGRSGKSCRLRWFNQL & DPRINKRAFTEEEEFRLLAAHRAYGNKWALISRLFPGRTDNAVKNHWHVIMARRT | REETRKGPWTEQEDLQLVCTVRLFGERRWDFIAKVSGLNRTGKSCRLRWVNYL & HPGLKRGRMSPHEERLILELHARWGNRWSRIARRLPGRTDNEIKNYWRTHMRKKA | 52 & 55 | 53 & 55 | 45.80% | 37.70% |
| 3 | MYB62 | HTH DNA-binding domain | 2 | 2 | DEELRRGPWTLEEDTLLTNYILHNGEGRWNHVAKCAGLKRTGKSCRLRWLNYL & KPDIRRGNLTPQEQLLILELHSKWGNRWSKIAQYLPGRTDNEIKNYWRTRVQKQA | MVELRRGPWTLEEDNLLMNYIACHGEGRWNLLARCSGLKRTGKSCRLRWLNYL & KPDIKRGNLTPEEQLLILELHSKWGNRWSRIAQHLPGRTDNEIKNYWRTRVQKQA | 53 & 55 | 53 & 55 | 82.40% | 92.70% |
| 4 | MYB81 | HTH DNA-binding domain and SANT domain | 2 | 2 | KKSFTKGPWTQAEDNLLIAYVDKHGDGNWNAVQNNSGLSRCGKSCRLRWVNHL & RPDLKKGAFTEKEEKRVIELHALLGNKWARMAEELPGRTDNEIKNFWNTRLKRLQ | GAARLWTAAENKQFERALAGLDLCRPDWEKVARAIPGRTVREVVSHFKSLQVDVQ & QDRKKGVPWTEEEHRLFLLGLKKYGKGDWRNISRNYVQTRTPTQVASHAQKYFIRLN | 53 & 55 | 55 & 57 | 66.70% | 44.40% |
| 5 | MYB88 | HTH DNA-binding domain | 2 | 2 | KKERHIVTWSPEEDDILRKQISLQGTENWAIIASKFNDKSTRQCRRRWYTYL & NSDFKRGGWSPEEDTLLCEAQRLFGNRWTEIAKVVSGRTDNAVKNRFTTLCKKRA | REEMRKGPWTEQEDIQLVCTVRLFGDHRWDFIAQVSGLNRTGKSCRLRWVNYL & HPGLKHGRMSPQEERIIIELHARWGNRWSRIARRLPGRTDNEIKNYWRTHMRKKA | 52 & 55 | 53 & 55 | 33.30% | 43.10% |
| 6 | WRKY25 | WRKY domain | 2 | 1 | NDGYGWRKYGQKQVKKSENPRSYFKCTYPDCVSKKIVETASDGQITEIIYKGGHNHPKP & SDIDVLIDGFRWRKYGQKVVKGNTNPRSYYKCTFQGCGVKKQVERSAADERAVLTTYEGRHNHDIP | GAEGPVDDGYSWRKYGQKDILGAKHPRAYYRCTHRNSQNCPATKQVQRTDDHPALFDVVYHGEH | 59 & 66 | 64 | 37.90% | NA |
Table S4: Performance evaluation details for the universal model of PTFSpot across various experimentally validated datasets.
Sheet 1: Performace of PTFSpot on Dataset 'E' TFs
| S.No. | TF | Sensitivity | Specificity | Accuracy | F1-Score | MCC |
|---|---|---|---|---|---|---|
| S.No. | TF | Sensitivity | Specificity | Accuracy | F1-Score | MCC |
| 1 | FUS3 | 96.52471855 | 98.43367597 | 97.47919726 | 97.45490487 | 95.08458941 |
| 2 | ABR1 | 99.09491194 | 98.8258317 | 98.96037182 | 98.96176866 | 97.94235273 |
| 3 | ARF2 | 98.42786219 | 98.90176019 | 98.66481119 | 98.66163996 | 97.36524739 |
| 4 | AT1G20910 | 95.89004537 | 98.61222311 | 97.25113424 | 97.21320346 | 94.65141852 |
| 5 | BPC1 | 95.79386681 | 99.16530575 | 97.47958628 | 97.43637068 | 95.08344341 |
| 6 | BAM8 | 99.09394373 | 98.37863615 | 98.73628994 | 98.74079354 | 97.50445533 |
| 7 | bHLH10 | 97.19309742 | 98.98427261 | 98.08868502 | 98.07141283 | 96.24983218 |
| 8 | ABF2 | 95.94553707 | 99.00151286 | 97.47352496 | 97.43432171 | 95.07242089 |
| 9 | At1g78700 | 98.99787047 | 99.37366905 | 99.18576976 | 99.18423695 | 98.38478754 |
| 10 | Adof1 | 98.69981752 | 98.43521898 | 98.56751825 | 98.56941091 | 97.17606669 |
| 11 | GATA11 | 96.99453552 | 99.10746812 | 98.05100182 | 98.03019146 | 96.17712456 |
| 12 | CRC | 96.82557743 | 98.21367619 | 97.51962681 | 97.50229148 | 95.16183296 |
| 13 | AT2G15740 | 98.73652291 | 98.14690027 | 98.44171159 | 98.4462921 | 96.93193512 |
| 14 | AT3G12130 | 95.92663173 | 99.4203589 | 97.67349532 | 97.63213189 | 95.45248552 |
| 15 | AT2G20110 | 97.79728387 | 98.59224909 | 98.19476648 | 98.18756235 | 96.45459834 |
| 16 | DEL2 | 97.99676898 | 99.03069467 | 98.51373183 | 98.50600844 | 97.07148712 |
| 17 | EIN3 | 98.33448994 | 99.16724497 | 98.75086745 | 98.7456446 | 97.53285606 |
| 18 | At1g25550 | 95.95626294 | 98.4989648 | 97.22761387 | 97.19191323 | 94.60721188 |
| 19 | AtGRF6 | 98.07292869 | 98.92684053 | 98.49988461 | 98.49345231 | 97.04466846 |
| 20 | ANL2 | 96.18552232 | 98.87299523 | 97.52925878 | 97.49560633 | 95.17887398 |
| 21 | ATHB13 | 95.9777815 | 98.72159884 | 97.34969017 | 97.31282458 | 94.83792676 |
| 22 | HSF3 | 96.82854816 | 98.895337 | 97.86194258 | 97.8396173 | 95.81441917 |
| 23 | LBD13 | 96.97580645 | 98.91129032 | 97.94354839 | 97.92345277 | 95.97092366 |
| 24 | AGL15 | 97.66392355 | 98.56649854 | 98.11521104 | 98.10666667 | 96.3013201 |
| 25 | AT5G23930 | 98.83320074 | 99.00556881 | 98.91938478 | 98.91845266 | 97.86212097 |
| 26 | MYB101 | 96.97477509 | 98.53589698 | 97.75533604 | 97.73767723 | 95.61090828 |
| 27 | At1g19000 | 98.47704295 | 98.40298796 | 98.44001545 | 98.44059286 | 96.9287011 |
| 28 | ANAC004 | 97.0071912 | 98.61463621 | 97.81091371 | 97.79317697 | 95.71711686 |
| 29 | FRS9 | 96.10510046 | 99.38176198 | 97.74343122 | 97.70584538 | 95.58634892 |
| 30 | AT1G24250 | 96.52087475 | 98.87342611 | 97.69715043 | 97.66974015 | 95.49912134 |
| 31 | RAV1 | 98.2081292 | 98.53928507 | 98.37370713 | 98.37100988 | 96.80029329 |
| 32 | REM19 | 98.85783718 | 98.68772783 | 98.7727825 | 98.77382542 | 97.57568275 |
| 33 | NLP7 | 98.55222337 | 98.65563599 | 98.60392968 | 98.60320745 | 97.24683813 |
| 34 | AT3G09735 | 97.52219296 | 98.50746269 | 98.01482782 | 98.00499975 | 96.10828503 |
| 35 | SPL15 | 97.90999237 | 98.30663616 | 98.10831426 | 98.10455518 | 96.28816883 |
| 36 | SRS7 | 96.4737243 | 98.4843869 | 97.4790556 | 97.45345438 | 95.08422269 |
| 37 | At1g69690 | 96.14825581 | 99.41860465 | 97.78343023 | 97.74658293 | 95.66281843 |
| 38 | AT1G76880 | 97.76536313 | 98.69646182 | 98.23091248 | 98.22263798 | 96.52426779 |
| 39 | WRKY14 | 95.61929355 | 99.16997141 | 97.39463248 | 97.34754237 | 94.92184154 |
| 40 | AT3G42860 | 95.82450832 | 98.36611195 | 97.09531014 | 97.05792216 | 94.35754722 |
| 41 | ATHB23 | 96.79906314 | 99.19000683 | 97.99453499 | 97.97027014 | 96.06838792 |
| 42 | ABI5 | 94.9656121 | 92.14580468 | 93.55570839 | 93.64530349 | 87.93720587 |
| 43 | AGL63 | 95.92009988 | 95.43570537 | 95.67790262 | 95.68834533 | 91.72931873 |
| 44 | AIL7 | 93.52777778 | 92.55555556 | 93.04166667 | 93.07532827 | 87.05108951 |
| 45 | ANAC005 | 95.68902661 | 94.89609916 | 95.29256289 | 95.31115247 | 91.02804305 |
| 46 | ANAC016 | 94.3174985 | 93.3674083 | 93.8424534 | 93.87156622 | 88.44269287 |
| 47 | ANAC017 | 95.84057777 | 93.01859034 | 94.42958406 | 94.50708869 | 89.47557526 |
| 48 | ANAC020 | 92.66840342 | 92.84207915 | 92.75524129 | 92.74894462 | 86.56019288 |
| 49 | ANAC028 | 93.62199645 | 97.41170779 | 95.51685212 | 95.43026218 | 91.42954572 |
| 50 | ANAC034 | 95.07131537 | 97.6703645 | 96.37083994 | 96.32305716 | 93.00273789 |
| 51 | ANAC038 | 97.32547597 | 94.96826836 | 96.14687217 | 96.19175627 | 92.58862075 |
| 52 | ANAC042 | 94.64849173 | 95.77180361 | 95.21014767 | 95.18309323 | 90.87857375 |
| 53 | ANAC045 | 96.96621961 | 94.26952593 | 95.61787277 | 95.67617295 | 91.61676417 |
| 54 | ANAC046 | 93.11735623 | 97.85757655 | 95.48746639 | 95.37791795 | 91.3725569 |
| 55 | ANAC047 | 92.21126525 | 93.9160956 | 93.06368043 | 93.00404585 | 87.08773592 |
| 56 | ANAC050 | 94.56301225 | 95.39884768 | 94.98092997 | 94.95986636 | 90.46534822 |
| 57 | ANAC053 | 95.01938924 | 92.97140087 | 93.99539506 | 94.05625862 | 88.70952992 |
| 58 | ANAC057 | 95.37553819 | 93.78089619 | 94.57821719 | 94.62110426 | 89.74304557 |
| 59 | ANAC058 | 94.25497534 | 97.65266202 | 95.95381868 | 95.88389246 | 92.23059979 |
| 60 | ANAC062 | 94.08857256 | 96.07552392 | 95.08204824 | 95.03269932 | 90.64597676 |
| 61 | ANAC070 | 92.01984869 | 93.67526234 | 92.84755552 | 92.78786015 | 86.71644098 |
| 62 | ANAC071 | 92.09131192 | 95.83794904 | 93.96463048 | 93.84941034 | 88.64983023 |
| 63 | ANAC075 | 97.25251678 | 93.37248322 | 95.3125 | 95.40170764 | 91.05774781 |
| 64 | ANAC083 | 97.1369886 | 96.16448692 | 96.65073776 | 96.66694477 | 93.52552061 |
| 65 | ANAC087 | 95.92671108 | 96.38098122 | 96.15384615 | 96.1450903 | 92.60347398 |
| 66 | ANAC094 | 93.14553991 | 95.58685446 | 94.36619718 | 94.29657795 | 89.36402349 |
| 67 | ANAC096 | 91.99801193 | 97.9787939 | 94.98840292 | 94.83391683 | 90.46221467 |
| 68 | ANAC103 | 97.15025907 | 93.23186528 | 95.19106218 | 95.28346832 | 90.83763494 |
| 69 | AREB3 | 95.58656036 | 93.67881549 | 94.63268793 | 94.68340149 | 89.8396892 |
| 70 | AT1G01250 | 93.14641745 | 98.33852544 | 95.74247144 | 95.62899787 | 91.83655391 |
| 71 | AT1G12630 | 96.45400705 | 93.09543444 | 94.77472074 | 94.86101895 | 90.08993741 |
| 72 | AT1G19040 | 92.25 | 92.41666667 | 92.33333333 | 92.32693912 | 85.84220256 |
| 73 | At1g19210 | 94.77520114 | 95.90156176 | 95.33838145 | 95.31197944 | 91.11081295 |
| 74 | At1g22810 | 92.88823039 | 96.32373368 | 94.60598204 | 94.51170679 | 89.78786178 |
| 75 | AT1G28160 | 95.6841521 | 94.76824868 | 95.22620039 | 95.24796243 | 90.90780275 |
| 76 | At1g36060 | 95.20710059 | 93.58974359 | 94.39842209 | 94.44335746 | 89.42301506 |
| 77 | AT1G44830 | 92.62428688 | 93.92828036 | 93.27628362 | 93.23215751 | 87.45566827 |
| 78 | AT1G47655 | 93.62820909 | 98.26012991 | 95.9441695 | 95.84801108 | 92.20902969 |
| 79 | At1g49010 | 95.81486478 | 94.19926768 | 95.00706623 | 95.04707588 | 90.51148266 |
| 80 | AT1G49560 | 97.43349156 | 92.76962137 | 95.10155646 | 95.21318195 | 90.6729176 |
| 81 | At1g64620 | 96.1661163 | 95.4720994 | 95.81910785 | 95.83356573 | 91.98761995 |
| 82 | At1g68670 | 91.76733781 | 94.7395334 | 93.2534356 | 93.15166261 | 87.41164211 |
| 83 | AT1G69570 | 91.70844582 | 94.28130682 | 92.99487632 | 92.90358588 | 86.96687884 |
| 84 | AT1G71450 | 93.30757868 | 92.59175094 | 92.94966481 | 92.97480894 | 86.89313837 |
| 85 | At1g72010 | 91.66666667 | 93.66554054 | 92.6661036 | 92.59206597 | 86.40521376 |
| 86 | AT1G72740 | 91.78500986 | 96.78500986 | 94.28500986 | 94.13847165 | 89.20982403 |
| 87 | At1g74840 | 96.48270501 | 92.72250291 | 94.60260396 | 94.70220781 | 89.78064342 |
| 88 | AT1G77200 | 97.28423204 | 95.433108 | 96.35867002 | 96.39206371 | 92.98132456 |
| 89 | At2g03500 | 96.15895212 | 92.14532259 | 94.15213736 | 94.26718435 | 88.97937687 |
| 90 | AT2G20400 | 94.64690138 | 93.49392629 | 94.07041384 | 94.10440123 | 88.84328615 |
| 91 | AT2G28810 | 91.80627283 | 95.57406474 | 93.69016879 | 93.56901573 | 88.16824087 |
| 92 | AT2G33550 | 95.72941578 | 96.44687393 | 96.08814486 | 96.07406137 | 92.48214847 |
| 93 | At2g33710 | 96.11955364 | 93.60174957 | 94.8606516 | 94.92454659 | 90.24647423 |
| 94 | AT2G40260 | 94.82358403 | 93.8718663 | 94.34772516 | 94.37449463 | 89.33393158 |
| 95 | At2g44940 | 95.06462985 | 92.12690952 | 93.59576968 | 93.68847713 | 88.00665563 |
| 96 | At2g45680 | 95.13761468 | 93.48623853 | 94.31192661 | 94.35850773 | 89.26947449 |
| 97 | At3g04030 | 94.54407504 | 98.46530556 | 96.5046903 | 96.43479026 | 93.24856146 |
| 98 | At3g09600 | 93.27287917 | 96.29442685 | 94.78365301 | 94.70363694 | 90.1070049 |
| 99 | AT3G10030 | 92.35023041 | 96.6359447 | 94.49308756 | 94.37249823 | 89.58316814 |
| 100 | AT3G10113 | 97.41915818 | 94.48268855 | 95.95092336 | 96.00951308 | 92.2264042 |
| 101 | At3g11280 | 94.03520209 | 91.98174707 | 93.00847458 | 93.07952896 | 86.99183708 |
| 102 | At3g12730 | 95.14859171 | 93.60168552 | 94.37513861 | 94.41831036 | 89.38178875 |
| 103 | At3g14180 | 94.90975301 | 92.72482584 | 93.81728942 | 93.88410337 | 88.39632983 |
| 104 | AT3G16280 | 92.85027015 | 95.69980016 | 94.27503516 | 94.19228892 | 89.2011979 |
| 105 | At3g24120 | 91.96359983 | 94.17656444 | 93.07008214 | 92.9925459 | 87.0974828 |
| 106 | AT3G25990 | 95.81739962 | 96.74952199 | 96.2834608 | 96.26605835 | 92.84286404 |
| 107 | At3g45610 | 94.93479357 | 93.24158801 | 94.08819079 | 94.13782017 | 88.87377735 |
| 108 | AT3G52440 | 92.09581244 | 93.24434205 | 92.67007725 | 92.62774096 | 86.41381394 |
| 109 | AT3G60490 | 93.2823412 | 94.06939364 | 93.67586742 | 93.65088198 | 88.15126095 |
| 110 | At3g60580 | 95.52134968 | 96.66057918 | 96.09096443 | 96.06857043 | 92.48705271 |
| 111 | At4g01280 | 93.04796534 | 95.18387076 | 94.11591805 | 94.0524005 | 88.92175982 |
| 112 | At4g16750 | 95.72586948 | 95.16412661 | 95.44499805 | 95.45775591 | 91.30481977 |
| 113 | AT4G18450 | 94.71788715 | 95.55822329 | 95.13805522 | 95.11754069 | 90.74855399 |
| 114 | At4g18890 | 93.62416107 | 95.97315436 | 94.79865772 | 94.73684211 | 90.13567665 |
| 115 | AT4G26030 | 93.20780094 | 95.06837032 | 94.13808563 | 94.08304107 | 88.96150279 |
| 116 | At4g31060 | 97.0546805 | 97.16993213 | 97.11230631 | 97.1106413 | 94.3913844 |
| 117 | At4g32800 | 93.75637105 | 96.76350663 | 95.25993884 | 95.18758085 | 90.96516554 |
| 118 | At4g36780 | 95.56456567 | 96.05602654 | 95.81029611 | 95.79997537 | 91.97156763 |
| 119 | AT4G37180 | 93.12607945 | 96.42733777 | 94.77670861 | 94.68904443 | 90.09368796 |
| 120 | AT5G02460 | 94.51292921 | 92.77320899 | 93.6430691 | 93.69788865 | 88.09254864 |
| 121 | At5g04390 | 93.0198915 | 94.17721519 | 93.59855335 | 93.56129502 | 88.0158747 |
| 122 | AT5G05550 | 92.26918306 | 93.7533227 | 93.01125288 | 92.95900377 | 86.99792605 |
| 123 | At5g05790 | 93.96195203 | 94.10669975 | 94.03432589 | 94.03000517 | 88.78042538 |
| 124 | At5g08330 | 93.50858369 | 96.19098712 | 94.84978541 | 94.77977162 | 90.22655473 |
| 125 | At5g08520 | 92.62523872 | 95.16674012 | 93.89598942 | 93.8174243 | 88.53345912 |
| 126 | At5g08750 | 97.19557196 | 92.17712177 | 94.68634686 | 94.81641469 | 89.92477616 |
| 127 | At5g18450 | 96.73479561 | 93.51944167 | 95.12711864 | 95.20421931 | 90.72435406 |
| 128 | AT5G22990 | 94.54180154 | 96.95278491 | 95.74729322 | 95.69540165 | 91.85393304 |
| 129 | At5g29000 | 95.90364725 | 96.74741426 | 96.32553076 | 96.3099631 | 92.92084406 |
| 130 | AT5G45580 | 94.4860096 | 93.0480474 | 93.7670285 | 93.81152248 | 88.30984754 |
| 131 | At5g47390 | 94.99477413 | 92.33538497 | 93.66507955 | 93.74820927 | 88.12859101 |
| 132 | AT5G47660 | 94.06178307 | 93.66163368 | 93.86170837 | 93.87396502 | 88.47689697 |
| 133 | At5g52660 | 94.18816138 | 93.02662037 | 93.60739087 | 93.64430289 | 88.03128358 |
| 134 | AT5G56840 | 96.62425998 | 96.95175715 | 96.78800857 | 96.78274035 | 93.78232157 |
| 135 | At5g58900 | 93.88907677 | 93.22624281 | 93.55765979 | 93.57894027 | 87.94512973 |
| 136 | AT5G61620 | 94.08004447 | 95.23809524 | 94.65906985 | 94.6279644 | 89.88797255 |
| 137 | At5g62940 | 96.76543619 | 96.01059427 | 96.38801523 | 96.40159636 | 93.0367608 |
| 138 | AT5G63260 | 95.25659093 | 97.21582511 | 96.23620802 | 96.19897251 | 92.7543497 |
| 139 | At5g65130 | 93.17893401 | 92.98857868 | 93.08375635 | 93.09033281 | 87.12417789 |
| 140 | At5g66730 | 92.00605857 | 92.59508583 | 92.3005722 | 92.27782935 | 85.78652161 |
| 141 | AT5G66940 | 93.1277533 | 95.63656388 | 94.38215859 | 94.31079288 | 89.3921861 |
| 142 | ATAF1 | 92.17141726 | 93.18091845 | 92.67616785 | 92.63901323 | 86.42441443 |
| 143 | ATHB18 | 92.95401403 | 97.52143414 | 95.23772408 | 95.12642578 | 90.91961167 |
| 144 | ATHB20 | 96.88619733 | 92.29371688 | 94.5899571 | 94.71139615 | 89.75453115 |
| 145 | ATHB21 | 92.7662181 | 94.60576941 | 93.68599376 | 93.62737995 | 88.16732022 |
| 146 | ATHB24 | 95.87681604 | 93.19623491 | 94.53652548 | 94.60878344 | 89.66633518 |
| 147 | ATHB25 | 97.45175319 | 92.80375102 | 95.12775211 | 95.23841148 | 90.7203093 |
| 148 | AtHB32 | 93.85074627 | 93.43283582 | 93.64179104 | 93.65504915 | 88.09201453 |
| 149 | ATHB33 | 97.41375242 | 97.81490396 | 97.61432819 | 97.60953349 | 95.34244751 |
| 150 | ATHB34 | 96.79525722 | 95.77911927 | 96.28718824 | 96.30595653 | 92.84970689 |
| 151 | ATHB5 | 92.2714874 | 93.80649302 | 93.03899021 | 92.98515105 | 87.04556799 |
| 152 | ATHB6 | 94.93069691 | 94.41698168 | 94.67383929 | 94.68748489 | 89.91490528 |
| 153 | AtIDD11 | 93.38619223 | 96.77045059 | 95.07832141 | 94.99360677 | 90.63575356 |
| 154 | BBX31 | 95.64034648 | 92.80907724 | 94.22471186 | 94.30532762 | 89.11214605 |
| 155 | bHLH122 | 94.58963012 | 94.02187526 | 94.30575269 | 94.32187162 | 89.25982134 |
| 156 | bHLH28 | 95.45501075 | 92.67069301 | 94.06285188 | 94.14437153 | 88.82637385 |
| 157 | bHLH80 | 92.31724627 | 93.91412349 | 93.11568488 | 93.06027544 | 87.1776115 |
| 158 | BIM1 | 96.83830172 | 95.75429088 | 96.2962963 | 96.31626235 | 92.86652206 |
| 159 | BIM2 | 95.13069426 | 94.62109322 | 94.87589374 | 94.88891681 | 90.27679053 |
| 160 | bZIP16 | 95.65352055 | 93.2892835 | 94.47140202 | 94.53599306 | 89.55119522 |
| 161 | bZIP18 | 94.66292135 | 98.28785447 | 96.47538791 | 96.41032627 | 93.19478268 |
| 162 | bZIP28 | 95.42772861 | 95.9439528 | 95.68584071 | 95.67467652 | 91.74381082 |
| 163 | bZIP3 | 93.37592138 | 96.9041769 | 95.14004914 | 95.05277375 | 90.74673882 |
| 164 | bZIP44 | 93.37947638 | 98.19440265 | 95.78693951 | 95.68300956 | 91.91957179 |
| 165 | bZIP48 | 97.2365161 | 98.09414904 | 97.66533257 | 97.65527802 | 95.43951093 |
| 166 | bZIP52 | 95.90991457 | 94.8615066 | 95.38571059 | 95.40977274 | 91.19677084 |
| 167 | bZIP53 | 93.9556609 | 93.19576129 | 93.5757111 | 93.60002778 | 87.97650485 |
| 168 | bZIP68 | 93.53272784 | 94.95772001 | 94.24522393 | 94.20392714 | 89.15169584 |
| 169 | BZR1 | 91.90093708 | 92.03480589 | 91.96787149 | 91.96249163 | 85.2260315 |
| 170 | CBF1 | 96.73429683 | 97.69316287 | 97.21372985 | 97.2003072 | 94.58247678 |
| 171 | CBF2 | 93.50619014 | 96.97889901 | 95.24254458 | 95.15847861 | 90.93230565 |
| 172 | CBF3 | 91.82464455 | 93.40442338 | 92.61453397 | 92.55573248 | 86.3182635 |
| 173 | CBF4 | 93.20614607 | 96.91116641 | 95.05865624 | 94.96538957 | 90.59921909 |
| 174 | CDF3 | 96.52986848 | 96.88480431 | 96.7073364 | 96.70148259 | 93.63146535 |
| 175 | CEJ1 | 92.9439195 | 95.28776537 | 94.11584243 | 94.04606693 | 88.92111127 |
| 176 | CRF10 | 97.45606866 | 96.98610544 | 97.22108705 | 97.22760167 | 94.59656157 |
| 177 | CRF4 | 93.45613185 | 93.23800291 | 93.34706738 | 93.35431546 | 87.57933546 |
| 178 | CUC1 | 92.9725667 | 95.96016535 | 94.46636603 | 94.38245112 | 89.54049513 |
| 179 | CUC2 | 94.00798935 | 96.25261556 | 95.13030245 | 95.07502886 | 90.73255224 |
| 180 | CUC3 | 95.65044687 | 96.90168818 | 96.27606753 | 96.25262316 | 92.82892745 |
| 181 | DAG2 | 91.73589025 | 93.55389946 | 92.64489486 | 92.5774233 | 86.36948978 |
| 182 | DDF1 | 92.54892122 | 92.24786754 | 92.39839438 | 92.40981964 | 85.95241326 |
| 183 | DDF2 | 95.95744681 | 98.29787234 | 97.12765957 | 97.09364909 | 94.41880091 |
| 184 | DEAR2 | 91.75035576 | 97.01158772 | 94.38097174 | 94.22916319 | 89.37879874 |
| 185 | DEAR3 | 95.54858403 | 93.24232661 | 94.39545532 | 94.45934619 | 89.41631758 |
| 186 | DREB19 | 95.06825939 | 92.05631399 | 93.56228669 | 93.65779851 | 87.94799881 |
| 187 | E2FA | 94.93606999 | 94.65006729 | 94.79306864 | 94.80050399 | 90.12833959 |
| 188 | ERF105 | 95.72592345 | 94.30867091 | 95.01729718 | 95.05235747 | 90.5301903 |
| 189 | ERF10 | 92.96402056 | 94.33466591 | 93.64934323 | 93.60552041 | 88.10418627 |
| 190 | ERF115 | 95.57318856 | 95.48966381 | 95.53142619 | 95.53329159 | 91.46221243 |
| 191 | ERF11 | 94.38190853 | 96.19714025 | 95.28952439 | 95.24637978 | 91.02134236 |
| 192 | ERF13 | 92.56480787 | 96.41577061 | 94.49028924 | 94.38211797 | 89.580015 |
| 193 | ERF15 | 94.92107421 | 95.59758098 | 95.25932759 | 95.24323768 | 90.96792803 |
| 194 | ERF2 | 92.62883235 | 96.0208741 | 94.32485323 | 94.22694094 | 89.2877043 |
| 195 | ERF38 | 92.74098167 | 93.79065642 | 93.26581904 | 93.23028907 | 87.43793003 |
| 196 | ERF3 | 96.06732481 | 98.18740897 | 97.12736689 | 97.09658952 | 94.41852221 |
| 197 | ERF4 | 97.48345284 | 92.15388858 | 94.81867071 | 94.95315805 | 90.16038159 |
| 198 | ERF5 | 93.45535219 | 96.06211869 | 94.75873544 | 94.68951953 | 90.06351713 |
| 199 | ERF7 | 97.29411162 | 94.08086916 | 95.68749039 | 95.75568053 | 91.74268499 |
| 200 | ERF8 | 96.75069628 | 92.41591088 | 94.58330358 | 94.6982141 | 89.74382294 |
| 201 | ESE1 | 95.61068702 | 92.36641221 | 93.98854962 | 94.08450704 | 88.69391223 |
| 202 | ESE3 | 96.93746923 | 96.29739045 | 96.61742984 | 96.62822086 | 93.46356141 |
| 203 | GATA19 | 97.05882353 | 96.60633484 | 96.83257919 | 96.83972912 | 93.86574667 |
| 204 | GATA1 | 91.64179104 | 96.11940299 | 93.88059701 | 93.74045802 | 88.49865087 |
| 205 | GATA20 | 97.36821884 | 98.15583218 | 97.76202551 | 97.75317737 | 95.62408594 |
| 206 | GATA4 | 92.24868793 | 92.69277352 | 92.47073072 | 92.45397532 | 86.07512206 |
| 207 | GBF3 | 95.97636112 | 97.38463473 | 96.68049793 | 96.65695834 | 93.58074165 |
| 208 | GBF6 | 91.68302028 | 97.75721895 | 94.72011961 | 94.55474171 | 89.97944951 |
| 209 | GT2 | 95.30524506 | 95.03869304 | 95.17196905 | 95.17839509 | 90.8101031 |
| 210 | GTL1 | 91.75902442 | 95.8169753 | 93.78799986 | 93.65934962 | 88.33820689 |
| 211 | HAT2 | 93.8238342 | 93.78238342 | 93.80310881 | 93.80439287 | 88.37424583 |
| 212 | HDG1 | 94.38940157 | 93.31370662 | 93.8515541 | 93.88444645 | 88.45850834 |
| 213 | HSFA6B | 94.9622814 | 92.42484855 | 93.69356498 | 93.77257336 | 88.17875139 |
| 214 | HSFB3 | 94.83418367 | 94.19642857 | 94.51530612 | 94.53273999 | 89.63203875 |
| 215 | HSFC1 | 92.89871944 | 92.61415082 | 92.75643513 | 92.76672694 | 86.56220049 |
| 216 | HY5 | 97.10610932 | 92.6759557 | 94.89103251 | 95.00174764 | 90.2946155 |
| 217 | IDD4 | 96.70745619 | 92.39863113 | 94.55304366 | 94.66791869 | 89.68994185 |
| 218 | IDD5 | 95.45981625 | 95.81660869 | 95.63821247 | 95.6304173 | 91.65687565 |
| 219 | JKD | 97.12817412 | 97.52116082 | 97.32466747 | 97.31940027 | 94.79244282 |
| 220 | LBD18 | 92.91761 | 97.95255439 | 95.43508219 | 95.31719385 | 91.27594927 |
| 221 | LBD19 | 96.06299213 | 92.71077957 | 94.38688585 | 94.47941669 | 89.3979699 |
| 222 | LBD23 | 97.45715865 | 93.14538419 | 95.30127142 | 95.4004329 | 91.03581037 |
| 223 | LBD2 | 95.97851282 | 92.88421208 | 94.43136245 | 94.516205 | 89.47789234 |
| 224 | LCL1 | 93.42228464 | 92.74344569 | 93.08286517 | 93.10626385 | 87.12236873 |
| 225 | LEP | 96.07609988 | 92.15219976 | 94.11414982 | 94.22740525 | 88.91265507 |
| 226 | LOB | 93.56417012 | 94.0158073 | 93.78998871 | 93.77593361 | 88.35114342 |
| 227 | MGP | 94.5491388 | 93.41438703 | 93.98176292 | 94.0157163 | 88.68718122 |
| 228 | MP | 91.77169929 | 93.95056762 | 92.86113346 | 92.78250359 | 86.73838972 |
| 229 | MYB107 | 92.18768467 | 93.71823662 | 92.95296064 | 92.89861545 | 86.89760248 |
| 230 | MYB116 | 96.24478442 | 93.08066759 | 94.66272601 | 94.74584973 | 89.89013234 |
| 231 | MYB118 | 93.72215472 | 95.3017416 | 94.51194816 | 94.46825883 | 89.62497694 |
| 232 | MYB119 | 93.87604752 | 92.21843632 | 93.04724192 | 93.1043931 | 87.05952375 |
| 233 | MYB121 | 97.28711379 | 93.82064808 | 95.55388093 | 95.62962963 | 91.49802974 |
| 234 | MYB13 | 95.30201342 | 93.43773304 | 94.36987323 | 94.42186923 | 89.37186743 |
| 235 | MYB27 | 92.49278152 | 93.33012512 | 92.91145332 | 92.8816508 | 86.82739478 |
| 236 | MYB33 | 93.13548524 | 97.04024209 | 95.08786367 | 94.99005049 | 90.65120825 |
| 237 | MYB44 | 94.77056277 | 97.6969697 | 96.23376623 | 96.17784026 | 92.74812611 |
| 238 | MYB49 | 91.69839431 | 95.10842576 | 93.40341003 | 93.28898619 | 87.669966 |
| 239 | MYB55 | 92.92920583 | 97.6804346 | 95.30482021 | 95.19056662 | 91.04048006 |
| 240 | MYB56 | 93.70092791 | 98.14418273 | 95.92255532 | 95.82991149 | 92.16993743 |
| 241 | MYB57 | 94.74476987 | 96.20641562 | 95.47559275 | 95.44228392 | 91.35966827 |
| 242 | MYB58 | 96.46127682 | 96.94531659 | 96.7032967 | 96.69529867 | 93.62388377 |
| 243 | MYB61 | 96.47038267 | 95.90541092 | 96.18789679 | 96.19863511 | 92.66631917 |
| 244 | MYB62 | 93.69564295 | 96.18174876 | 94.93869586 | 94.87498926 | 90.38676118 |
| 245 | MYB63 | 95.19355677 | 96.54455703 | 95.8690569 | 95.84096259 | 92.0786851 |
| 246 | MYB65 | 93.27968874 | 97.70978866 | 95.4947387 | 95.39268436 | 91.38701721 |
| 247 | MYB67 | 92.53045404 | 96.56146179 | 94.54595792 | 94.43377034 | 89.67849124 |
| 248 | MYB70 | 93.009167 | 93.63889996 | 93.32403348 | 93.3029467 | 87.53919048 |
| 249 | MYB73 | 96.65375832 | 98.2126899 | 97.43322411 | 97.41305979 | 94.99760783 |
| 250 | MYB74 | 93.16770186 | 93.76663709 | 93.46716948 | 93.447547 | 87.78767742 |
| 251 | MYB77 | 96.43697 | 98.47146013 | 97.45421506 | 97.42805205 | 95.03702546 |
| 252 | MYB81 | 92.02014846 | 95.44008484 | 93.73011665 | 93.62103844 | 88.23959968 |
| 253 | MYB83 | 92.25821273 | 92.18355157 | 92.22088215 | 92.22378506 | 85.65205379 |
| 254 | MYB88 | 92.54744175 | 92.81167427 | 92.67955801 | 92.66987372 | 86.43084607 |
| 255 | MYB93 | 94.6969697 | 96.10125647 | 95.39911308 | 95.36657983 | 91.22072417 |
| 256 | MYB96 | 93.30035971 | 92.8057554 | 93.05305755 | 93.07019511 | 87.07115716 |
| 257 | MYB98 | 93.59952146 | 94.88559892 | 94.24256019 | 94.20529801 | 89.14718543 |
| 258 | MYB99 | 92.25873298 | 94.22735346 | 93.24304322 | 93.17587264 | 87.39677521 |
| 259 | NAM | 96.14285714 | 94.21428571 | 95.17857143 | 95.22461974 | 90.82036077 |
| 260 | NAP | 96.83421943 | 97.86128565 | 97.34775254 | 97.33406206 | 94.83592117 |
| 261 | NGA4 | 93.82239382 | 95.84942085 | 94.83590734 | 94.78303267 | 90.20316096 |
| 262 | NST1 | 97.17630854 | 95.41580579 | 96.29605716 | 96.32837668 | 92.86539365 |
| 263 | NTL8 | 93.75259731 | 97.54813686 | 95.65036709 | 95.56622423 | 91.67314622 |
| 264 | NTM1 | 92.84432762 | 96.49100847 | 94.66766804 | 94.56863578 | 89.89731384 |
| 265 | OBP1 | 94.09170889 | 95.56507176 | 94.82839032 | 94.79000929 | 90.1906274 |
| 266 | OBP3 | 94.18797131 | 95.83869781 | 95.01333456 | 94.97183392 | 90.52271537 |
| 267 | OBP4 | 92.44778819 | 94.91448775 | 93.68113797 | 93.6022311 | 88.15723744 |
| 268 | PHV | 93.12990171 | 95.49642111 | 94.31316141 | 94.24506559 | 89.2701244 |
| 269 | PLT3 | 96.47435897 | 92.1474359 | 94.31089744 | 94.43137255 | 89.25909708 |
| 270 | PUCHI | 94.86700887 | 93.72375175 | 94.29538031 | 94.3278042 | 89.2409114 |
| 271 | RKD2 | 92.48400853 | 97.47044001 | 94.97722427 | 94.84879358 | 90.44720777 |
| 272 | RRTF1 | 95.07676019 | 98.18687136 | 96.63181578 | 96.57861128 | 93.48738603 |
| 273 | RVE1 | 93.44709465 | 97.00900787 | 95.22805126 | 95.1415239 | 90.90578169 |
| 274 | SGR5 | 91.81333841 | 92.3461136 | 92.079726 | 92.05857102 | 85.4138598 |
| 275 | SMB | 96.87477854 | 97.86927456 | 97.37202655 | 97.3588937 | 94.88192501 |
| 276 | SPL1 | 95.54177166 | 95.9972863 | 95.76952898 | 95.75987178 | 91.8969116 |
| 277 | SPL5 | 95.57893825 | 95.84307612 | 95.71100719 | 95.70533527 | 91.78989492 |
| 278 | SPL9 | 95.82841701 | 93.69695124 | 94.76268412 | 94.81791135 | 90.07170484 |
| 279 | STOP1 | 97.2541507 | 96.80715198 | 97.03065134 | 97.03727302 | 94.23758575 |
| 280 | STZ | 96.5891909 | 92.38018214 | 94.48468652 | 94.59836418 | 89.56854181 |
| 281 | SVP | 96.10403549 | 95.03434459 | 95.56919004 | 95.59276195 | 91.53053721 |
| 282 | TCP20 | 96.02649007 | 95.28145695 | 95.65397351 | 95.67010309 | 91.68547522 |
| 283 | TCP24 | 93.31671953 | 96.7829633 | 95.04984141 | 94.96253602 | 90.58412407 |
| 284 | TCP3 | 96.86126805 | 95.10357815 | 95.9824231 | 96.01742377 | 92.28647425 |
| 285 | TCP7 | 93.57249626 | 92.52615845 | 93.04932735 | 93.08550186 | 87.06418372 |
| 286 | TCX2 | 96.44706857 | 94.99689248 | 95.72198053 | 95.75277664 | 91.8091293 |
| 287 | TGA1 | 92.92670425 | 97.28344439 | 95.10507432 | 94.99607021 | 90.68055076 |
| 288 | TGA3 | 96.42072903 | 97.10144928 | 96.76108915 | 96.75002754 | 93.73184397 |
| 289 | TGA4 | 96.6493672 | 98.27005372 | 97.45971046 | 97.43895722 | 95.04783279 |
| 290 | TGA5 | 95.54582904 | 94.67044284 | 95.10813594 | 95.12945399 | 90.69452209 |
| 291 | TGA6 | 95.35280578 | 94.65830621 | 95.005556 | 95.02283918 | 90.50977257 |
| 292 | TGA9 | 94.75606193 | 97.44376278 | 96.09991236 | 96.04678709 | 92.501336 |
| 293 | TINY | 97.34513274 | 93.06784661 | 95.20648968 | 95.30685921 | 90.86421519 |
| 294 | TRP2 | 94.36579023 | 97.44293557 | 95.9043629 | 95.84036388 | 92.14049999 |
| 295 | VIP1 | 94.00544959 | 96.89373297 | 95.44959128 | 95.38291402 | 91.30969009 |
| 296 | VND1 | 93.54263289 | 98.34998216 | 95.94630753 | 95.84647019 | 92.21232625 |
| 297 | VND4 | 95.4409979 | 95.97637708 | 95.70868749 | 95.69716926 | 91.78556453 |
| 298 | VND6 | 91.7216429 | 94.70525644 | 93.21344967 | 93.11067424 | 87.34241945 |
| 299 | VRN1 | 94.39977025 | 94.83055715 | 94.6151637 | 94.60354008 | 89.81016209 |
| 300 | WIP5 | 96.18939823 | 96.53838898 | 96.3638936 | 96.35753768 | 92.99216993 |
| 301 | WRKY15 | 96.29250307 | 92.39297419 | 94.34273863 | 94.45093238 | 89.31747312 |
| 302 | WRKY18 | 97.32966277 | 92.50516173 | 94.91741225 | 95.03712913 | 90.34029785 |
| 303 | WRKY20 | 92.761004 | 98.36304111 | 95.56202255 | 95.43413174 | 91.50474055 |
| 304 | WRKY21 | 95.62251279 | 96.98692439 | 96.30471859 | 96.279336 | 92.88187711 |
| 305 | WRKY22 | 93.37804748 | 95.8107101 | 94.59437879 | 94.52781894 | 89.77014939 |
| 306 | WRKY24 | 92.39130435 | 93.87871854 | 93.13501144 | 93.08357349 | 87.21117007 |
| 307 | WRKY25 | 92.79642224 | 97.19101124 | 94.99371674 | 94.88124256 | 90.47953998 |
| 308 | WRKY27 | 97.34984411 | 93.32901935 | 95.33943173 | 95.43128181 | 91.1061218 |
| 309 | WRKY28 | 95.18282355 | 95.32230746 | 95.25256551 | 95.24925224 | 90.9558849 |
| 310 | WRKY29 | 96.48465091 | 93.98533007 | 95.23499049 | 95.29380199 | 90.92125617 |
| 311 | WRKY30 | 93.26783115 | 96.77947598 | 95.02365357 | 94.93471618 | 90.53676997 |
| 312 | WRKY31 | 92.53398707 | 95.76554491 | 94.14976599 | 94.05368671 | 88.97829391 |
| 313 | WRKY33 | 95.2890305 | 92.01183432 | 93.65043241 | 93.75279893 | 88.10082783 |
| 314 | WRKY3 | 95.92457421 | 92.57907543 | 94.25182482 | 94.34639545 | 89.15842682 |
| 315 | WRKY40 | 91.99417758 | 95.97282872 | 93.98350315 | 93.86138614 | 88.68204082 |
| 316 | WRKY45 | 96.19016393 | 95.37704918 | 95.78360656 | 95.80067921 | 91.92250561 |
| 317 | WRKY50 | 92.27323214 | 92.62865605 | 92.45094409 | 92.43750464 | 86.04156493 |
| 318 | WRKY55 | 95.22073714 | 96.43580397 | 95.82827055 | 95.80277099 | 92.00401763 |
| 319 | WRKY65 | 91.92149414 | 96.47356758 | 94.19753086 | 94.06238865 | 89.05714499 |
| 320 | WRKY6 | 94.16510112 | 96.28688253 | 95.22599182 | 95.17480174 | 90.90576194 |
| 321 | WRKY70 | 93.80482456 | 93.95833333 | 93.88157895 | 93.87687918 | 88.51184588 |
| 322 | WRKY71 | 95.22905937 | 92.55218216 | 93.89062076 | 93.97131106 | 88.52362571 |
| 323 | WRKY75 | 93.1020562 | 92.80062439 | 92.95134029 | 92.96194776 | 86.89629313 |
| 324 | WRKY7 | 91.91358025 | 98.27160494 | 95.09259259 | 94.93146318 | 90.64812143 |
| 325 | WRKY8 | 92.70531699 | 96.23251802 | 94.4689175 | 94.36962009 | 89.54320509 |
Sheet 2: Performace of PTFSpot on Dataset 'D' TFs
| S.No. | TF | Sensitivity | Specificity | Accuracy | F1-Score | MCC |
|---|---|---|---|---|---|---|
| S.No. | TF | Sensitivity | Specificity | Accuracy | F1-Score | MCC |
| 1 | ABF1 | 94.88134816 | 92.78917804 | 93.8352631 | 93.89908387 | 88.42807532 |
| 2 | ABF3 | 91.40069456 | 95.75822722 | 93.57946089 | 93.43645662 | 87.97200848 |
| 3 | ABF4 | 95.22901721 | 96.03672302 | 95.63287011 | 95.61516182 | 91.64690427 |
| 4 | ABI5 | 90.9047619 | 93.0952381 | 92 | 91.91141069 | 85.27647014 |
| 5 | ANAC032 | 93.56201839 | 95.37658464 | 94.46930152 | 94.41866299 | 89.54865613 |
| 6 | ANAC102 | 96.11054248 | 96.51995906 | 96.31525077 | 96.30769231 | 92.90198959 |
| 7 | AP1 | 96.20746207 | 92.59942599 | 94.40344403 | 94.5026178 | 89.42645352 |
| 8 | AT5G04760 | 94.39878234 | 96.12959339 | 95.26418787 | 95.22284611 | 90.97558336 |
| 9 | AZF1 | 92.51743817 | 94.54660748 | 93.53202283 | 93.4657271 | 87.89825063 |
| 10 | BBM | 93.49566238 | 95.11336491 | 94.30451364 | 94.25806997 | 89.25639356 |
| 11 | BZIP28 | 91.07711918 | 93.37157425 | 92.22434672 | 92.13410703 | 85.65413588 |
| 12 | CCA1 | 95.68690096 | 96.80511182 | 96.24600639 | 96.2248996 | 92.77341052 |
| 13 | CRY2 | 95.6484274 | 96.72554933 | 96.18698837 | 96.16634178 | 92.66433251 |
| 14 | DELLA | 93.66906475 | 91.79856115 | 92.73381295 | 92.80114041 | 86.52121874 |
| 15 | DREB2A | 92.47537204 | 95.26304758 | 93.86920981 | 93.78254862 | 88.4856841 |
| 16 | ERF115 | 90.46385898 | 93.4714461 | 91.96765254 | 91.84501845 | 85.21900077 |
| 17 | FBH3 | 94.08425456 | 92.68001195 | 93.38213325 | 93.42827474 | 87.6389714 |
| 18 | FHY1 | 96.15384615 | 93.69230769 | 94.92307692 | 94.98480243 | 90.35874005 |
| 19 | FHY3 | 91.7989418 | 91.26984127 | 91.53439153 | 91.55672823 | 84.50189668 |
| 20 | FIE | 92.77575758 | 91.56363636 | 92.16969697 | 92.21686747 | 85.56460663 |
| 21 | FLM | 93.49865952 | 93.43163539 | 93.46514745 | 93.46733668 | 87.78437812 |
| 22 | GBF2 | 95.13099202 | 95.60218634 | 95.36658918 | 95.35564722 | 91.16245018 |
| 23 | GBF3 | 94.20308483 | 92.85025707 | 93.52667095 | 93.57016326 | 87.89031393 |
| 24 | HAT22 | 94.55703925 | 91.81183488 | 93.18443706 | 93.27672094 | 87.29313025 |
| 25 | HB5 | 91.03377919 | 92.00861483 | 91.52119701 | 91.47966739 | 84.47945865 |
| 26 | HB6 | 92.39119053 | 94.25499983 | 93.32309518 | 93.26028749 | 87.53564792 |
| 27 | HB7 | 94.21626153 | 96.89857502 | 95.55741827 | 95.49702634 | 91.50651767 |
| 28 | HSFA1A | 95.89788348 | 92.49399956 | 94.19594152 | 94.29307016 | 89.05930172 |
| 29 | HSFA6A | 93.29451917 | 96.24573379 | 94.77012648 | 94.69179827 | 90.08297435 |
| 30 | IBH1 | 92.55474453 | 94.74452555 | 93.64963504 | 93.57933579 | 88.10296291 |
| 31 | JAG | 91.27801333 | 94.48818898 | 92.88310115 | 92.76700523 | 86.77240314 |
| 32 | KAN1 | 90.8718745 | 93.67577773 | 92.27382612 | 92.16396875 | 85.73592701 |
| 33 | LEC1 | 92.20992823 | 92.71830144 | 92.46411483 | 92.44491081 | 86.06384089 |
| 34 | LFY | 94.8968052 | 93.22872491 | 94.06276506 | 94.11187439 | 88.82899198 |
| 35 | MYB3 | 92.08326884 | 96.76520662 | 94.42423773 | 94.29058204 | 89.45875827 |
| 36 | MYB3R3 | 90.7578085 | 93.11315924 | 91.93548387 | 91.83937824 | 85.16758515 |
| 37 | MYB44 | 91.49080764 | 90.66099149 | 91.07589956 | 91.11277338 | 83.74403086 |
| 38 | NFYB2 | 90.83298255 | 94.20045835 | 92.51672045 | 92.38856382 | 86.14558884 |
| 39 | NFYC2 | 94.21161214 | 94.57707044 | 94.39434129 | 94.38407937 | 89.4170801 |
| 40 | PhyA | 94.55081001 | 92.63622975 | 93.59351988 | 93.65426696 | 88.00570267 |
| 41 | PIF1 | 92.80177187 | 93.35548173 | 93.0786268 | 93.05941144 | 87.11516423 |
| 42 | PIF4 | 90.52489386 | 90.78348128 | 90.65418757 | 90.64208838 | 83.05520269 |
| 43 | PIF5 | 91.68406402 | 93.94572025 | 92.81489214 | 92.7327116 | 86.65889056 |
| 44 | RD26 | 94.82924947 | 93.44973953 | 94.1394945 | 94.17964072 | 88.96484988 |
| 45 | REV | 90.46541694 | 95.6043956 | 93.03490627 | 92.85121911 | 87.02300358 |
| 46 | RGA | 92.36769977 | 96.45793801 | 94.41281889 | 94.29616875 | 89.44116847 |
| 47 | SEP3 | 93.99291572 | 92.48871852 | 93.24081712 | 93.29127336 | 87.39393973 |
| 48 | SOC1 | 91.92904656 | 94.63414634 | 93.28159645 | 93.18948078 | 87.46135007 |
| 49 | SVP | 92.89665855 | 92.45408276 | 92.67537066 | 92.69154339 | 86.42361226 |
| 50 | WRKY33 | 92.14001795 | 91.63567979 | 91.88784887 | 91.90825375 | 85.09164806 |
| 51 | ZAT6 | 93.1651557 | 93.44107213 | 93.30311391 | 93.29386225 | 87.50314592 |
Sheet 3: Performace of all tools on on common TFs between Arabidopsis thaliana and Zea mays
| Tool | Sensitivity | Specificity | Accuracy | F1-Score | MCC | TF |
|---|---|---|---|---|---|---|
| Tool | Sensitivity | Specificity | Accuracy | F1-Score | MCC | TF |
| k-mer grammar | 3.916410931 | 93.05442475 | 48.48541784 | 7.065381139 | 22.76404593 | LHY1 |
| k-mer grammar | 32.02328652 | 67.97671348 | 50 | 39.0417013 | 46.65659409 | MYB56 |
| k-mer grammar | 72.91608195 | 44.06399102 | 58.49003649 | 63.72332597 | 49.37932376 | MYB62 |
| k-mer grammar | 43.58542093 | 60.77784897 | 52.18163495 | 47.68448097 | 49.35213778 | MYB81 |
| k-mer grammar | 69.85388254 | 41.12954735 | 55.49171495 | 61.08128589 | 48.52259835 | MYB88 |
| k-mer grammar | 25.32538679 | 81.15376676 | 53.23957677 | 35.13227746 | 41.73549711 | WRKY25 |
| SeqConv | 0.676395065 | 98.55117847 | 49.61378677 | 1.324638688 | 10.26792219 | LHY1 |
| SeqConv | 80.85622372 | 19.14377628 | 50 | 61.79012463 | 39.34327716 | MYB56 |
| SeqConv | 13.93488633 | 91.4959304 | 52.71540836 | 22.76217765 | 31.7942224 | MYB62 |
| SeqConv | 19.5426467 | 82.46604998 | 51.00434834 | 28.51346936 | 38.88675141 | MYB81 |
| SeqConv | 2.57991722 | 98.08384379 | 50.33188051 | 4.93782615 | 14.83139733 | MYB88 |
| SeqConv | 0.005822293 | 99.99576561 | 50.00079395 | 0.011643415 | 0.709091231 | WRKY25 |
| TSPTFBS | 1.025030793 | 98.31736289 | 49.67119684 | 1.996016098 | 11.5657615 | LHY1 |
| TSPTFBS | 30.02320759 | 69.97679241 | 50 | 37.51812567 | 45.83587858 | MYB56 |
| TSPTFBS | 49.04574797 | 64.44007859 | 56.74291328 | 53.13568985 | 50.32429206 | MYB62 |
| TSPTFBS | 86.94316054 | 17.10720307 | 52.0251818 | 64.44149353 | 35.90195841 | MYB81 |
| TSPTFBS | 58.27112626 | 56.91743646 | 57.59428136 | 57.87937267 | 51.14898649 | MYB88 |
| TSPTFBS | 24.34406576 | 81.99005976 | 53.16706276 | 34.2021506 | 41.10177232 | WRKY25 |
| DNABERT | 16.82698709 | 99.99833577 | 58.41266143 | 28.80626781 | 30.30968552 | LHY1 |
| DNABERT | 70.19893854 | 67.97671348 | 69.08782601 | 69.42752066 | 57.27635471 | MYB56 |
| DNABERT | 95.91676565 | 21.47675208 | 58.69675887 | 69.89998609 | 35.6520726 | MYB62 |
| DNABERT | 94.11608799 | 14.90856083 | 54.51232441 | 67.41658361 | 31.18828455 | MYB81 |
| DNABERT | 98.08743169 | 16.29135891 | 57.1893953 | 69.61589686 | 30.56066945 | MYB88 |
| DNABERT | 96.36323317 | 26.18361237 | 61.27342277 | 71.33271776 | 39.18694076 | WRKY25 |
| PTFSpot | 92.77259347 | 92.94377988 | 92.85818668 | 92.85206853 | 86.73646387 | LHY1 |
| PTFSpot | 95.65981218 | 94.12503071 | 94.89242144 | 94.93131804 | 90.30544889 | MYB56 |
| PTFSpot | 93.08167275 | 92.59051361 | 92.83609318 | 92.85364317 | 86.69845714 | MYB62 |
| PTFSpot | 91.60876138 | 91.51482728 | 91.56179433 | 91.56575564 | 84.54764814 | MYB81 |
| PTFSpot | 91.97915582 | 91.7821941 | 91.88067496 | 91.88866307 | 85.07978976 | MYB88 |
| PTFSpot | 92.37385473 | 92.95767193 | 92.66576333 | 92.64429138 | 86.40711557 | WRKY25 |
| TSPTFBS 2.0 | 98.21959096 | 2.043057051 | 50.131324 | 66.32503235 | 13.69500182 | LHY1 |
| TSPTFBS 2.0 | 99.20969448 | 0.643141668 | 49.92641808 | 66.45733994 | 8.436201491 | MYB56 |
| TSPTFBS 2.0 | 98.93234165 | 1.115642994 | 50.02399232 | 66.43841134 | 10.39107873 | MYB62 |
| TSPTFBS 2.0 | 99.45399815 | 0.369564569 | 49.91178136 | 66.50563318 | 6.751616235 | MYB81 |
| TSPTFBS 2.0 | 98.99856503 | 1.21008838 | 50.10432671 | 66.48918464 | 10.45826122 | MYB88 |
| TSPTFBS 2.0 | 98.71352587 | 1.400308197 | 50.05691704 | 66.4037251 | 11.51264068 | WRKY25 |
| AgentBind | 1.5094064 | 91.6654514 | 46.5874289 | 2.748274 | 22.1709941 | LHY1 |
| AgentBind | 6.7458189 | 95.4775952 | 51.1117071 | 12.1253287 | 23.1115258 | MYB56 |
| AgentBind | 48.1694561 | 83.9696653 | 66.0695607 | 58.671763 | 52.2172598 | MYB62 |
| AgentBind | 2.7431623 | 97.5602848 | 50.1517236 | 5.2159865 | 15.8894418 | MYB81 |
| AgentBind | 46.0739702 | 77.956653 | 62.0153116 | 54.8116886 | 50.4369825 | MYB88 |
| AgentBind | 30.6677572 | 84.3270981 | 57.4974276 | 41.9128318 | 43.5243217 | WRKY25 |
Sheet 4: Performace of PTFSpot on inter-species TFs
| S.No. | TF | Sensitivity | Specificity | Accuracy | F1-Score | MCC | Species |
|---|---|---|---|---|---|---|---|
| S.No. | TF | Sensitivity | Specificity | Accuracy | F1-Score | MCC | Species |
| 1 | ALF2 | 95.40834993 | 93.85950648 | 94.63392821 | 94.67516489 | 89.84253325 | Maize |
| 2 | ALF7 | 95.42196182 | 89.8576393 | 92.63980056 | 92.83903029 | 86.34201174 | Maize |
| 3 | bHLH108 | 93.90953415 | 91.63559635 | 92.77256525 | 92.85381515 | 86.5863817 | Maize |
| 4 | bHLH118 | 95.40606517 | 90.92517653 | 93.16562085 | 93.31538591 | 87.2526632 | Maize |
| 5 | bHLH129 | 95.73159524 | 93.17182485 | 94.45171005 | 94.52182441 | 89.51566044 | Maize |
| 6 | bHLH136 | 94.23033649 | 93.03169865 | 93.63101757 | 93.66896068 | 88.07245727 | Maize |
| 7 | bHLH145 | 95.0214367 | 94.3039636 | 94.66270015 | 94.68177855 | 89.89487563 | Maize |
| 8 | bHLH162 | 95.18216254 | 90.90650453 | 93.04433353 | 93.18992137 | 87.04448723 | Maize |
| 9 | bHLH163 | 95.63920699 | 88.94220659 | 92.29070679 | 92.54048854 | 85.73831197 | Maize |
| 10 | bHLH172 | 96.49431979 | 93.82257608 | 95.15844793 | 95.22227225 | 90.78242433 | Maize |
| 11 | bHLH43 | 95.45487522 | 89.6156776 | 92.53527641 | 92.74703392 | 86.16152822 | Maize |
| 12 | bHLH47 | 95.30885914 | 88.73532234 | 92.02209074 | 92.27596198 | 85.28553006 | Maize |
| 13 | bHLH91 | 93.44392904 | 91.7743706 | 92.60914982 | 92.67033633 | 86.30888565 | Maize |
| 14 | bZIP104 | 95.15250981 | 92.5866624 | 93.8695861 | 93.94723842 | 88.48702667 | Maize |
| 15 | bZIP113 | 94.95467087 | 89.18013402 | 92.06740244 | 92.29001054 | 85.36905217 | Maize |
| 16 | bZIP38 | 94.69739819 | 92.10030166 | 93.39884992 | 93.48347021 | 87.66504858 | Maize |
| 17 | bZIP40 | 95.0866878 | 88.65031906 | 91.86850343 | 92.122031 | 85.02860349 | Maize |
| 18 | bZIP9 | 94.69095304 | 93.45032557 | 94.07063931 | 94.1071932 | 88.84356666 | Maize |
| 19 | C3H40 | 94.44617542 | 92.40535909 | 93.42576725 | 93.49217364 | 87.71338849 | Maize |
| 20 | COL13 | 95.39861393 | 89.75767291 | 92.57814342 | 92.78173252 | 86.23617686 | Maize |
| 21 | COL18 | 95.41765776 | 92.21701413 | 93.81733595 | 93.91472001 | 88.39324514 | Maize |
| 22 | COL2 | 94.97938956 | 90.47056644 | 92.724978 | 92.88537102 | 86.49479193 | Maize |
| 23 | COL3 | 95.78471961 | 93.16428303 | 94.47450132 | 94.54596113 | 89.55604486 | Maize |
| 24 | COL7 | 94.694268 | 88.7380424 | 91.7161552 | 91.95572285 | 84.77788424 | Maize |
| 25 | COL8 | 95.8632252 | 92.30739239 | 94.0853088 | 94.1886301 | 88.86326619 | Maize |
| 26 | DOF17 | 90.80732934 | 93.74009782 | 92.27371358 | 92.15873044 | 85.73521046 | Maize |
| 27 | DOF21 | 92.6074896 | 92.1220527 | 92.36477115 | 92.38325839 | 85.89531051 | Maize |
| 28 | DOF22 | 90.77877925 | 90.92109424 | 90.84993675 | 90.84342116 | 83.37432981 | Maize |
| 29 | DOF30 | 90.11870234 | 90.06204022 | 90.09037128 | 90.09317799 | 82.14475452 | Maize |
| 30 | EREB102 | 95.94723105 | 88.7319669 | 92.33959897 | 92.6063352 | 85.81620386 | Maize |
| 31 | EREB109 | 95.26546764 | 91.23516063 | 93.25031414 | 93.38364387 | 87.40159027 | Maize |
| 32 | EREB147 | 96.35450296 | 90.34517937 | 93.34984116 | 93.54382732 | 87.56185928 | Maize |
| 33 | EREB172 | 97.13521466 | 92.8246623 | 94.97993848 | 95.08585194 | 90.45506873 | Maize |
| 34 | EREB17 | 96.40335602 | 90.0791515 | 93.24125376 | 93.44842138 | 87.37104047 | Maize |
| 35 | EREB18 | 94.69940195 | 92.42052251 | 93.55996223 | 93.63251587 | 87.94627923 | Maize |
| 36 | EREB198 | 95.52667339 | 88.99640556 | 92.26153947 | 92.50622138 | 85.69043845 | Maize |
| 37 | EREB209 | 96.71782095 | 92.8291361 | 94.77347852 | 94.87316181 | 90.0858173 | Maize |
| 38 | EREB211 | 95.73796369 | 93.24623714 | 94.49210042 | 94.55987691 | 89.58771197 | Maize |
| 39 | EREB34 | 95.67494643 | 93.31005476 | 94.4925006 | 94.55686275 | 89.58874433 | Maize |
| 40 | EREB71 | 96.14074915 | 90.63822103 | 93.38948509 | 93.56648799 | 87.63432863 | Maize |
| 41 | EREB97 | 97.61647021 | 93.22428033 | 95.42037527 | 95.51878696 | 91.25181408 | Maize |
| 42 | EREB98 | 95.88538086 | 93.53381063 | 94.70959575 | 94.77107665 | 89.97619095 | Maize |
| 43 | GATA12 | 95.6928839 | 91.08926342 | 93.39107366 | 93.53977576 | 87.64266007 | Maize |
| 44 | GLK11 | 93.92182004 | 91.79910362 | 92.86046183 | 92.9354421 | 86.73739796 | Maize |
| 45 | GLK1 | 93.69756098 | 91.85365854 | 92.77560976 | 92.84160665 | 86.59277784 | Maize |
| 46 | GLK25 | 93.99812817 | 88.76269493 | 91.38041155 | 91.6002921 | 84.22522927 | Maize |
| 47 | GLK2 | 94.59343672 | 93.22251735 | 93.90797704 | 93.94945111 | 88.55713404 | Maize |
| 48 | GLK52 | 93.86892178 | 90.59972458 | 92.23432318 | 92.35921927 | 85.66711408 | Maize |
| 49 | GLK53 | 93.05838041 | 90.22222222 | 91.64030132 | 91.75719087 | 84.67213602 | Maize |
| 50 | GLK9 | 94.53987639 | 94.25382757 | 94.39685198 | 94.4048544 | 89.42156604 | Maize |
| 51 | GRAS23 | 94.55409797 | 88.93368636 | 91.74389217 | 91.96956394 | 84.82719193 | Maize |
| 52 | GRAS49 | 94.46137213 | 90.80499312 | 92.63318262 | 92.76544402 | 86.34265519 | Maize |
| 53 | HB33 | 95.47918206 | 93.28652377 | 94.38285292 | 94.44376752 | 89.39420575 | Maize |
| 54 | HB34 | 92.82782124 | 88.74215785 | 90.78498954 | 90.96946811 | 83.25436001 | Maize |
| 55 | HB38 | 95.27876854 | 92.26581566 | 93.7722921 | 93.86471866 | 88.31497644 | Maize |
| 56 | HB66 | 93.59044636 | 92.74158183 | 93.1660141 | 93.19489715 | 87.26563671 | Maize |
| 57 | HB70 | 94.62974984 | 90.40794927 | 92.51884955 | 92.67350457 | 86.14473805 | Maize |
| 58 | HB84 | 94.69591105 | 92.84889726 | 93.77240416 | 93.82939016 | 88.31847604 | Maize |
| 59 | KNOX6 | 93.4484062 | 92.18918427 | 92.81879523 | 92.863726 | 86.66792779 | Maize |
| 60 | LG2 | 95.50800077 | 94.34162329 | 94.92481203 | 94.95423834 | 90.36411948 | Maize |
| 61 | MYB31 | 94.80843841 | 90.23581474 | 92.52212658 | 92.6892726 | 86.1481908 | Maize |
| 62 | MYB37 | 94.01469538 | 93.42109739 | 93.71789638 | 93.73648643 | 88.22488185 | Maize |
| 63 | MYB38 | 94.58989015 | 93.56314539 | 94.07651777 | 94.10677198 | 88.854201 | Maize |
| 64 | MYB4 | 94.63915045 | 91.41722572 | 93.02818808 | 93.13872071 | 87.0217749 | Maize |
| 65 | MYB56 | 93.8547486 | 90.43330832 | 92.14402846 | 92.27616172 | 85.51391847 | Maize |
| 66 | MYB62 | 94.08362989 | 94.0688019 | 94.0762159 | 94.07665505 | 88.85425603 | Maize |
| 67 | MYB6 | 94.61373707 | 90.20545539 | 92.40959623 | 92.57329135 | 85.95787367 | Maize |
| 68 | MYB81 | 93.86126741 | 94.05843557 | 93.95985149 | 93.95389099 | 88.6493488 | Maize |
| 69 | MYB88 | 94.67841197 | 92.61855919 | 93.64848558 | 93.71323464 | 88.10128354 | Maize |
| 70 | MYBR17 | 93.55525704 | 94.21381999 | 93.88453851 | 93.86433491 | 88.51680542 | Maize |
| 71 | MYBR32 | 94.11883862 | 93.90501913 | 94.01192888 | 94.01832387 | 88.74097193 | Maize |
| 72 | MYBR40 | 97.40041145 | 90.95754629 | 94.17897887 | 94.36064683 | 89.01303187 | Maize |
| 73 | MYBR41 | 94.70211718 | 92.55046775 | 93.62629247 | 93.69413255 | 88.06230691 | Maize |
| 74 | MYBR43 | 93.8255641 | 90.18232366 | 92.00394388 | 92.14699579 | 85.27687367 | Maize |
| 75 | MYBR64 | 94.59235757 | 92.6748517 | 93.63360464 | 93.69406299 | 88.07563837 | Maize |
| 76 | MYBR87 | 94.21431017 | 91.51831565 | 92.86631291 | 92.96119579 | 86.74560447 | Maize |
| 77 | MYBR96 | 96.22871046 | 93.06569343 | 94.64720195 | 94.73053892 | 89.86239291 | Maize |
| 78 | MYBST1 | 93.53578976 | 90.78379123 | 92.1597905 | 92.26620743 | 85.54349052 | Maize |
| 79 | NAC109 | 92.77592352 | 90.36884087 | 91.57238219 | 91.67260582 | 84.56079 | Maize |
| 80 | NAC23 | 93.92260455 | 90.50053175 | 92.21156815 | 92.34258924 | 85.62792913 | Maize |
| 81 | NAC49 | 93.60670002 | 91.15727648 | 92.38198825 | 92.47415812 | 85.92043879 | Maize |
| 82 | SIG6 | 93.8354453 | 94.34588613 | 94.09066571 | 94.0755453 | 88.8795912 | Maize |
| 83 | TCP10 | 95.43490513 | 91.14784896 | 93.29137704 | 93.43216053 | 87.47139042 | Maize |
| 84 | TCP23 | 95.54229108 | 92.06248412 | 93.8023876 | 93.90837598 | 88.36595554 | Maize |
| 85 | WRKY25 | 94.63269739 | 92.01111292 | 93.32190516 | 93.40830854 | 87.53146987 | Maize |
| 86 | WRKY53 | 94.11208933 | 88.89413786 | 91.50311359 | 91.71915873 | 84.42904957 | Maize |
| 87 | WRKY82 | 93.8276935 | 91.39062988 | 92.60916169 | 92.69813721 | 86.30675061 | Maize |
| 88 | WRKY94 | 94.17137853 | 88.79217626 | 91.4817774 | 91.70488296 | 84.39226444 | Maize |
| 89 | WRKY95 | 94.67496191 | 91.2468258 | 92.96089385 | 93.07951567 | 86.90508838 | Maize |
| 90 | ZIM16 | 94.83242576 | 92.36793353 | 93.60017964 | 93.67808124 | 88.01587795 | Maize |
| 91 | ZIM18 | 94.90071204 | 88.71566936 | 91.8081907 | 92.05392478 | 84.92982657 | Maize |
| 92 | ZIM28 | 94.60269865 | 92.8577378 | 93.73021822 | 93.7844477 | 88.24485109 | Maize |
| 93 | ZIM3 | 94.56976458 | 94.11861209 | 94.34418834 | 94.35691779 | 89.32803218 | Maize |
| 94 | NAC10 | 92.20650636 | 89.46857951 | 90.83754294 | 90.96127971 | 83.3478627 | Rice |
| 95 | NAC5 | 94.9853612 | 88.8132362 | 91.8992987 | 92.14180743 | 85.08276557 | Rice |
| 96 | NAC6 | 95.19093891 | 90.68824658 | 92.93959275 | 93.09504714 | 86.86289995 | Rice |
| 97 | NAC9 | 97.02999003 | 90.51825677 | 93.7741234 | 93.9704379 | 88.29886851 | Rice |
| 98 | ARF4 | 94.17898434 | 93.16552027 | 93.6722523 | 93.70415537 | 88.14470371 | Rice |
| 99 | JMJ705 | 92.69079686 | 94.47250281 | 93.58164983 | 93.52395782 | 87.98529813 | Rice |
| 100 | OsABF1 | 94.58478904 | 94.18167159 | 94.38323031 | 94.39452863 | 89.39733652 | Rice |
| 101 | OsbZIP23 | 95.64905913 | 94.07775337 | 94.86340625 | 94.90344746 | 90.25330186 | Rice |
| 102 | OsbZIP39 | 93.43021756 | 92.91856565 | 93.17439161 | 93.19180873 | 87.28039533 | Rice |
| 103 | OsbZIP46 | 93.34435192 | 88.71654695 | 91.03044943 | 91.23330227 | 83.65249824 | Rice |
| 104 | OsGLK1 | 95.26670981 | 88.64056578 | 91.95363779 | 92.21167075 | 85.16980359 | Rice |
| 105 | OsGLK2 | 94.36528497 | 89.37823834 | 91.87176166 | 92.06951027 | 85.046359 | Rice |
| 106 | OSH1 | 93.27506353 | 91.67554718 | 92.47530535 | 92.53500724 | 86.08125148 | Rice |
| 107 | OsHOX24 | 92.11246201 | 88.86018237 | 90.48632219 | 90.63855242 | 82.77374689 | Rice |
| 108 | OsIDS1 | 93.44072538 | 93.01630173 | 93.22851355 | 93.24285302 | 87.37397396 | Rice |
| 109 | OsMADS29 | 94.8821595 | 93.10770074 | 93.99493012 | 94.04774031 | 88.70930107 | Rice |
| 110 | OsMYC2 | 91.79487179 | 94.1025641 | 92.94871795 | 92.86640726 | 86.88835928 | Rice |
| 111 | OsNAC5 | 91.49201597 | 89.52095808 | 90.50648703 | 90.59913527 | 82.81217259 | Rice |
| 112 | OsNAC6 | 93.51759793 | 92.66255717 | 93.09007755 | 93.11949312 | 87.13462543 | Rice |
| 113 | OsNF-YB1 | 97.6746824 | 89.4623412 | 93.5685118 | 93.82218348 | 87.92408447 | Rice |
| 114 | OsSRT1 | 93.87701928 | 90.12506514 | 92.00104221 | 92.1483376 | 85.27140523 | Rice |
| 115 | PROG1 | 92.68597625 | 91.21692019 | 91.95144822 | 92.01013601 | 85.1968833 | Rice |
| 116 | SDG711 | 96.71762763 | 89.71881842 | 93.21822302 | 93.44752078 | 87.32551666 | Rice |
| 117 | SNAC1 | 94.66783885 | 92.38208088 | 93.52495987 | 93.59812554 | 87.88528084 | Rice |
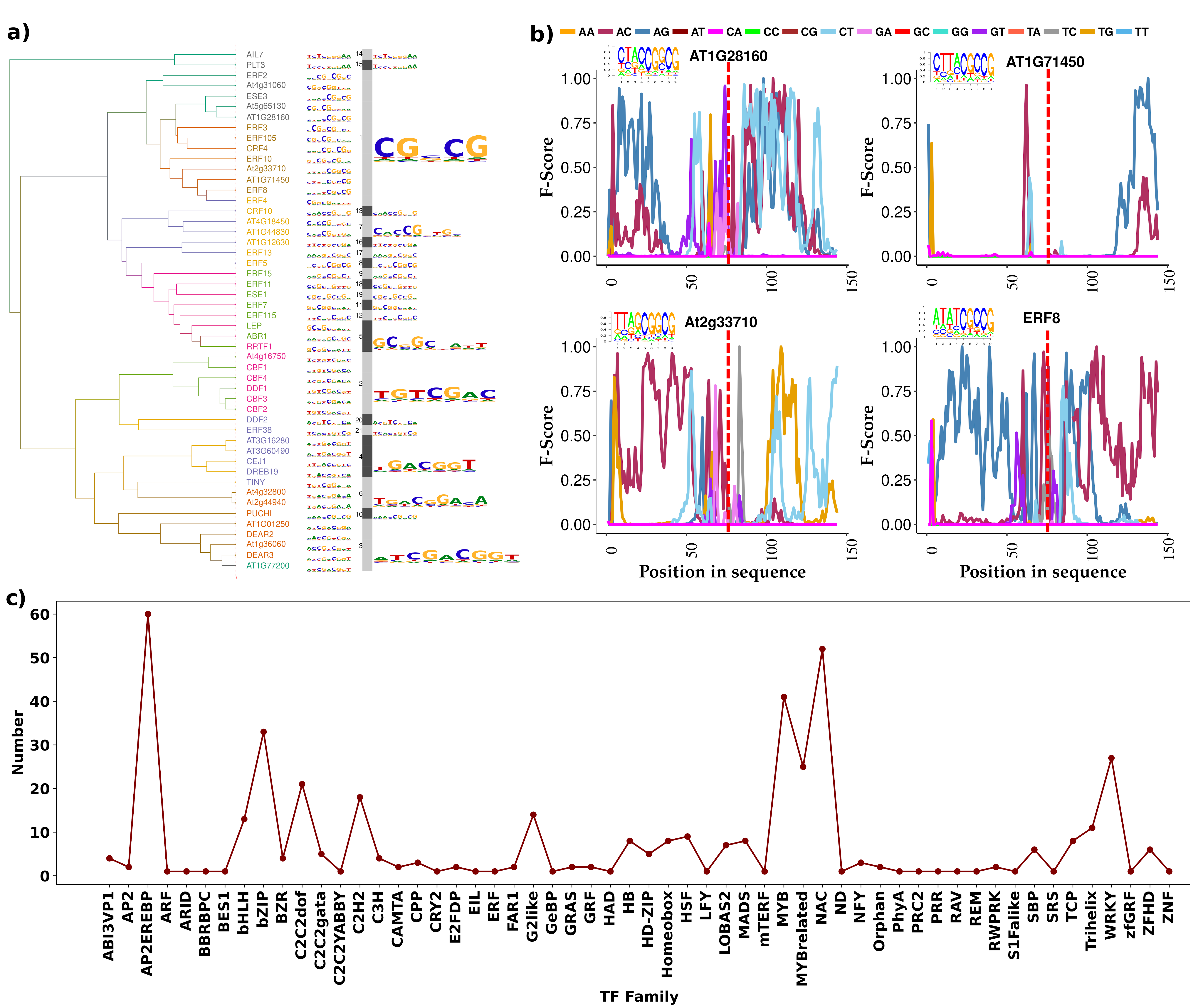
Figure-1: Motif and data. (a) Clustering plot of the prime motifs for TF family AP2EREBP. The TFs share similarity in their prime motifs. (b) Different characteristics spectra for TFs belonging to same family, same cluster, and sharing similar prime motif. This highlights the importance of context. Despite having similar binding motifs, their binding preferences differ from each other due to context. (c) Lineplot for the data abundance for the collected 441 TFs (ChIP and DAP-Seq) across 56 different family.
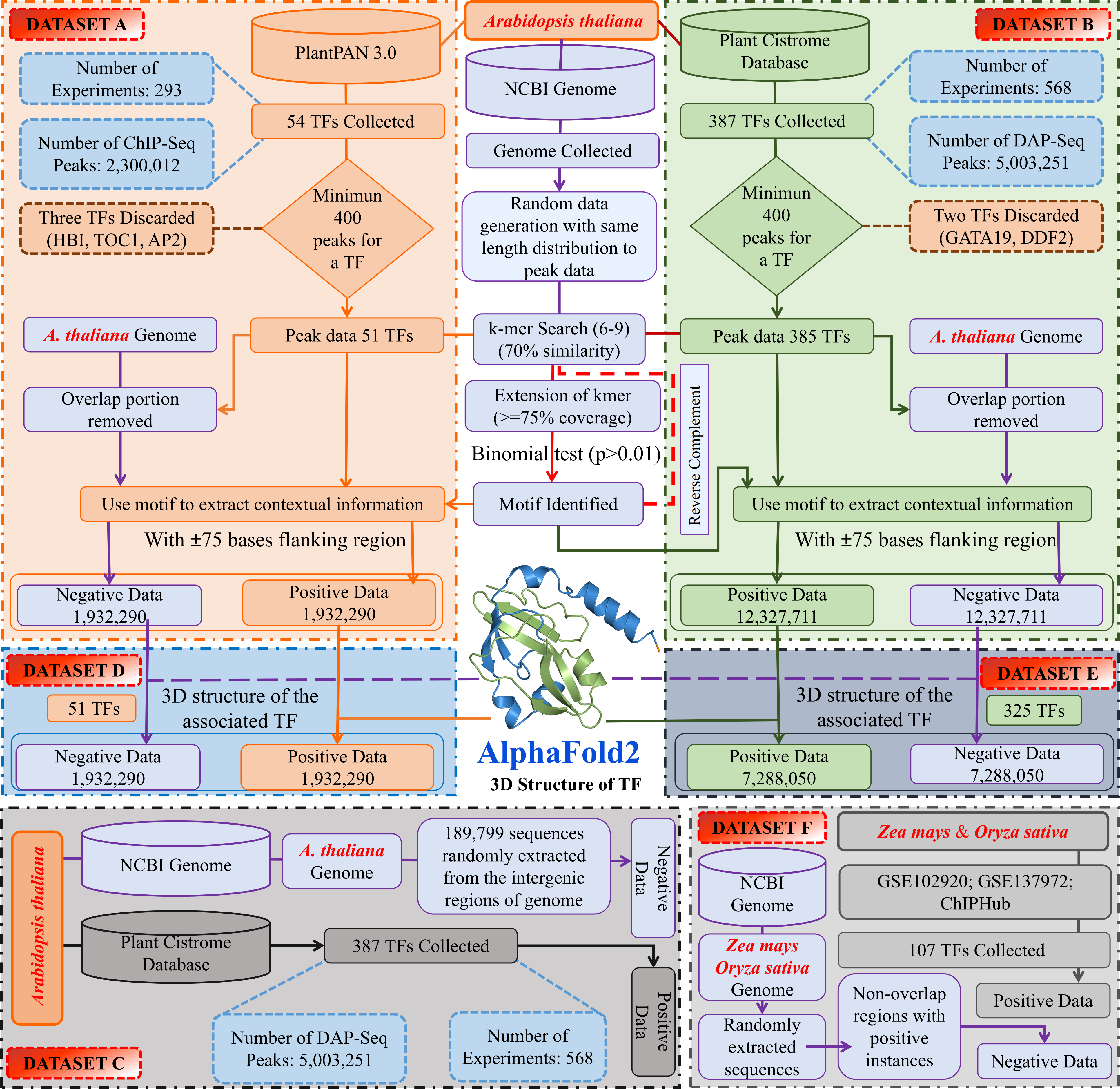
Figure-2: Flowchart representation of dataset formation. (a) The protocol followed for Dataset “A” creation, (b) Dataset “B” creation, (c) Dataset “C” creation, (d) Dataset "D" creation, (e) Dataset “E” creation, and (f) Dataset “F” creation. The datasets “A” and “B” contained the positive instances originating from ChIP-seq and DAP-seq, respectively, with sources PlantPAN3.0 and Plant Cistrome databases, respectively. Negative part of the datasets was formed by considering those genomic regions which display the regions similar to the prime motifs found enriched in DAP/ChIP-seq TF binding data but never appeared in the DAP/ChIP-seq data. Dataset “C” also contained the positive instances from the Plant Cistrome Database but for negative instances it contains random genomic regions. Most of the existing tools have used this dataset and its subsets. The datasets “D” and “E” were created from the datasets “A” and “B”, respectively, while adding up the TF structural data also. Dataset “F” was constructed to be used completely as a test set to evaluate the universal model and its applicability in cross-species manner. This dataset covered 117 TFs from Zea mays (93 TF) and Oryza sativa (24 TF).

Figure-3: Implementation of the PTFSpot Deep Co-learning system using Transformers and DenseNet to identify TFBR across plant genomes. The first part is a 14 heads attention transformers which learn from the dimeric, pentameric and heptameric words representations of any given sequence arising from anchoring prime motif’s context. In the parallel, the bound TF’s structure is learned by the DenseNet. Learning by both partners are joined finally together, which is passed on to the final fully connected layers to generate the probability score for existence of binding region in the center.
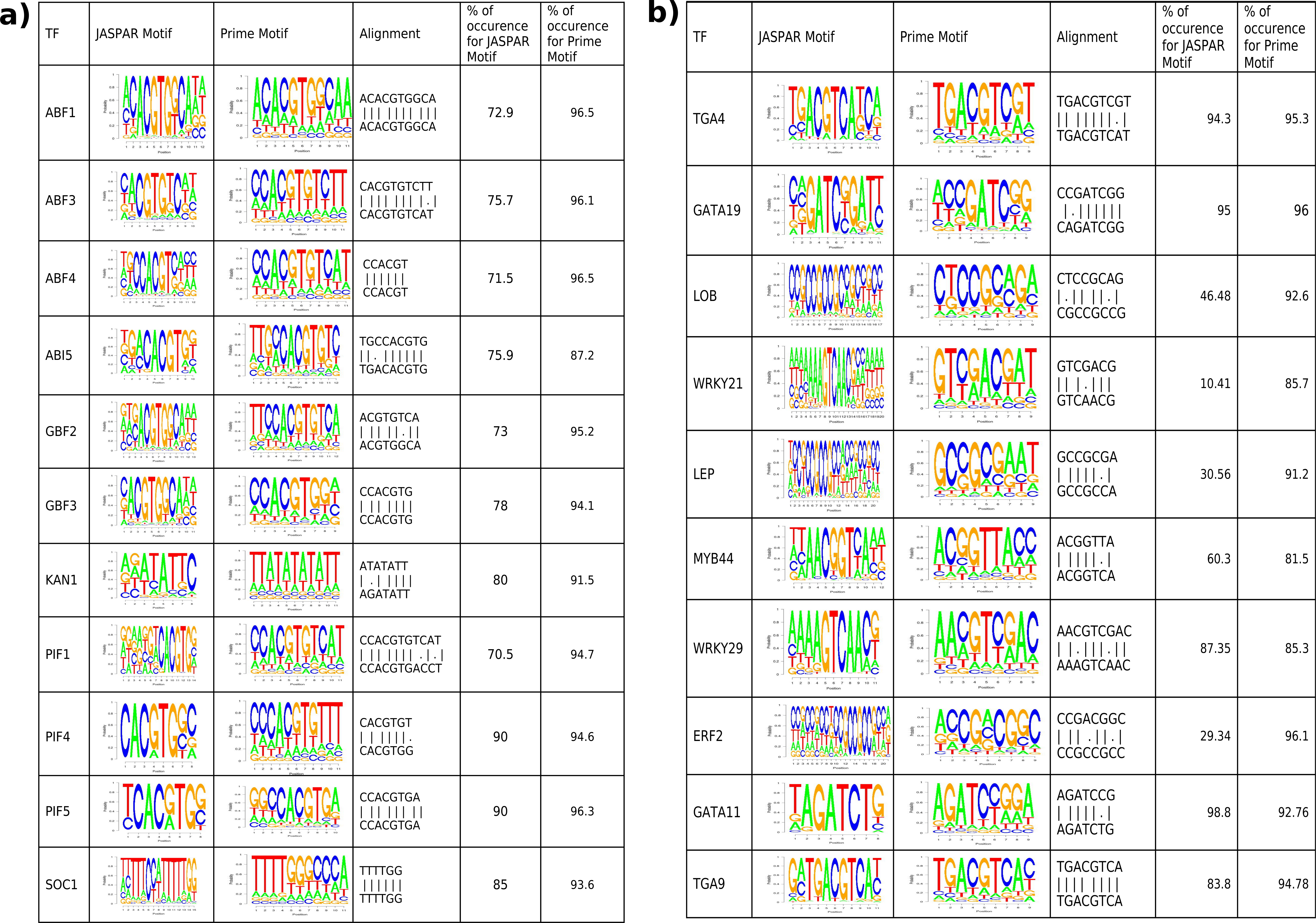
Supplementary Figure-S8: Comparison between experimentally reported motifs and motifs identified in the present study.(a) Motifs derives from ChIP-seq TF data and (b) Motifs derives from DAP-seq TF data. The similarity was determined using TOMTOM package. Several experimentally validated motifs were found similar to the identified prime motifs, with high significance (p-value less than 0.01).

Figure-4: (a) Ablation analysis for three main properties for discriminating between the negative and positive instances. These words representations appeared highly additive and complementary to each other as the performance increased substantially as they were combined together. (b) Ten-fold independent training-testing random trials on Dataset “B” depicts consistent performance of PTFSpot transformers. (c) Objective comparative benchmarking result on Dataset “B”. This datasets contained the TF originating from DAP-seq from the plant cistrome database. Here all the compared tools were train and tested on the Dataset “B”. (d) Performance dispersion plot on Dataset “B”. PTFSpot transformers consistently demonstrated minimal variance and distribution in its performance, maintaining a strong balance in identifying both positive and negative instances with a high level of precision. (e) Objective comparative benchmarking on Dataset ”C”. Here, all the compared tools were first trained and then tested Dataset ‘C’ and evaluated for their performance. This gave a clear view on the performance of each of the compared algorithms. (f) Performance dispersion plot on Dataset “C”. PTFSpot transformers consistently demonstrated high values across all the three performance metrics with least dispersion, affirming the algorithm's robustness. From the plots it is clearly visible that for all these datasets and associated benchmarkings, PTFSpot consistently and significantly outperformed the compared tools for all the compared metrics (MCC values were converted to percentage representation for scaling purpose).
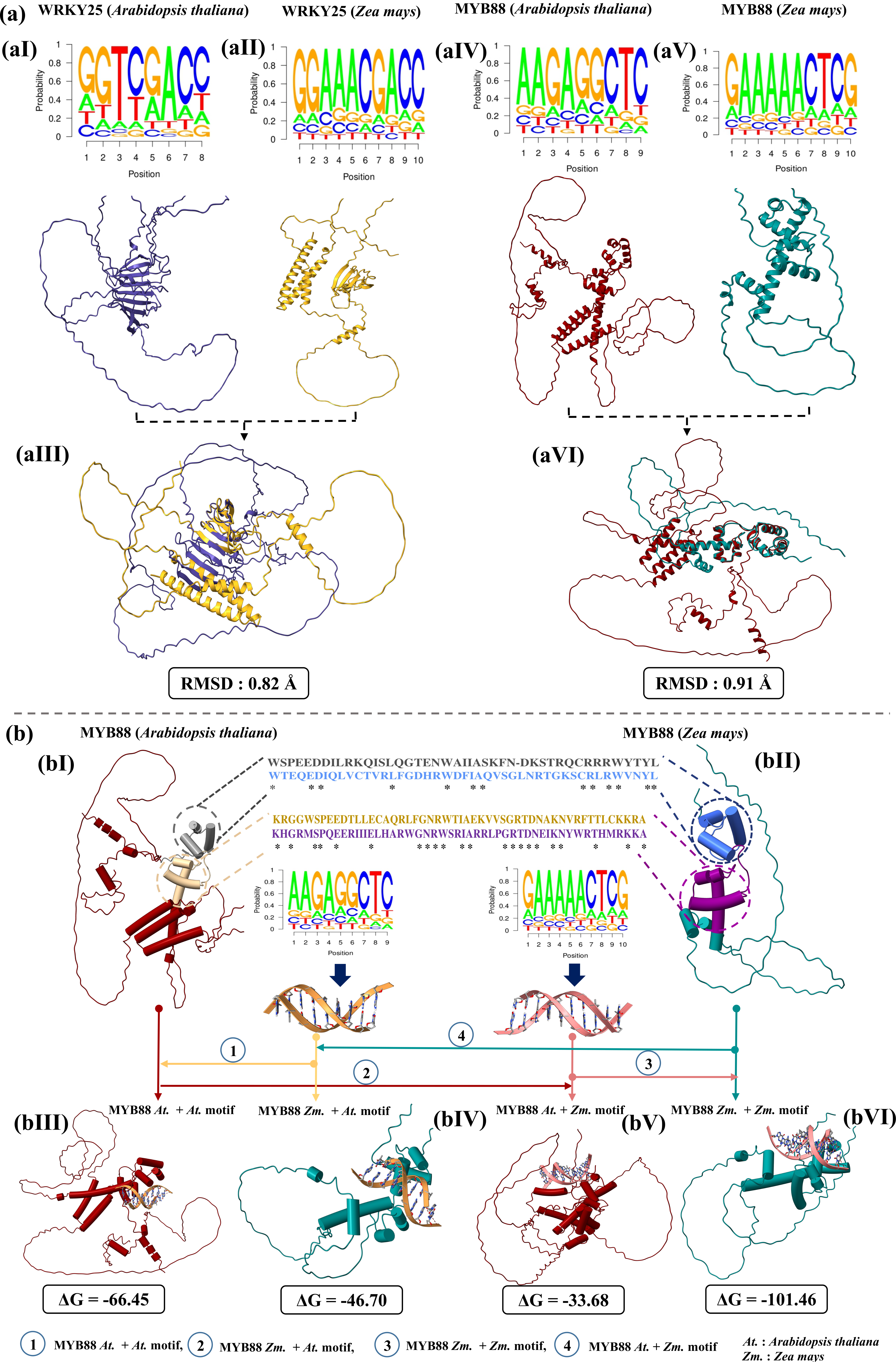
Figure-5: Co-variation in the structure of the transcription factor and the corresponding binding site across various species. (a) TF structure and its binding motif comparison across the species, (aI) prime binding motif of WRKY25 TF (Arabidopsis thaliana) and its 3D structure, (aII) prime binding motif of WRKY25 TF (Zea mays) and its 3D structure, (aIII) superimposed TF structures of Arabidopsis thaliana and Zea mays, with the structural differences measured in RMSD value. (aIV) prime binding motif of MYB88 TF (Arabidopsis thaliana) and its 3D structure, (aV) prime binding motif for WRKY25 TF (Zea mays) and its 3D structure, (aVI) superimposed TF structures for Arabidopsis thaliana and Zea mays, and corresponding structural difference in RMSD value. (b) Domain based comparison between Arabidopsis thaliana and Zea mays for MYB88, (bI) Arabidopsis thaliana’s MYB88’s has two domains. The first domain and its corresponding amino acids sequence are in grey color. The second domain and its corresponding amino acids sequence is shown in the tan color, (bII) Zea mays MYB88 too has two domains. The first domain and its corresponding amino acids sequence are in purple color. The second domain and its corresponding amino acids sequence is shown in maroon color, (bIII and bVI) The docking analysis shows the stability of the complexes when a TF was docked to its binding motif within the same species and to the one from another species for the same TF. It is clearly evident that the same binding motif can’t work across the species and it varies with species as well as the structure of the TF.

Figure-6: Performance and benchmarking of the universal model of PTFSpot. PTFSpot universal model was raised from 41 different TFs representing 41 different families, from A. thaliana. a) (i) The performance over the same 41 TF’s test set (Dataset “E”). (ii) The performance over the remain TF’s as test set from Dataset “E”. (iii) The complete dataset “D” (containing ChIP-seq TFs) worked as another test set. Performance on all of them was in the same range with exceptional accuracy. (b) Comparative benchmarking for the TFs whose models were available commonly among the compared tools. PTFSpot universal model outperformed them with huge leap for each TF. (c) Comparative benchmarking for the 13 wheat TFs (371,066 peak regions, equal number of negative instances following the same protocol as dataset “F”) whose models were available in TSPTFBS2.0 for comparison. PTFSpot universal model outperformed TSPTFBS2.0 for every compared TF by huge margins (p-value: 1.46e-05; kruskal wallis test). Reason to select TSPTFBS2.0 for this comparison was that among the existing software tools, TSPTFBS2.0 was found the best performer. (d) PTFSpot universal model was raised using A. thaliana TFs. It was tested in trans-species manner across rice and maize. For both, it returned outstanding results, reinforcing itself as the solution for reliable cross-species discovery of TFBR.

Figure-S1:Performance results for PTFSpot transformer for Datasets “A”.The developed approach demonstrated a consistently strong and dependable performance across the Dataset “A”.
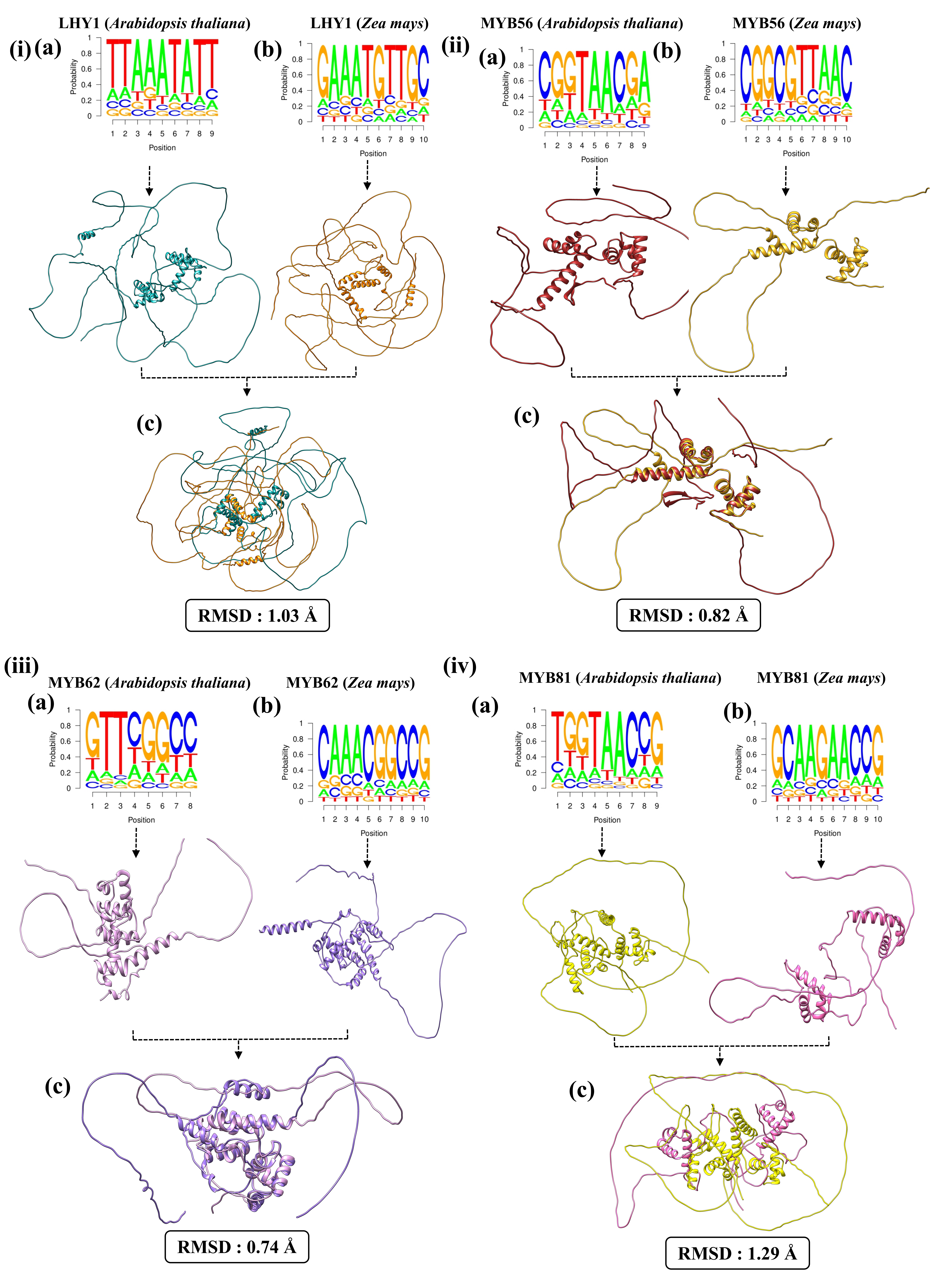
Figure-S2: 3D structure based co-variation in transcription factors and their binding sites across the species (A. thaliana vs Z. mays). (i) The prime binding motifs of LHY1 and their corresponding superimposed TF structures with the RMSD value indicating the structural variations. (ii) The prime binding motifs for MYB56 and their superimposed TF structures, with the structural differences expressed as the RMSD value. (iii) The prime binding motifs for MYB62, along with the comparison of their superimposed TF structures and structural differences expressed as the RMSD value. (iv) The prime binding motif for MYB81 and their superimposed TF structures with the structural differences measured in terms of the RMSD value.
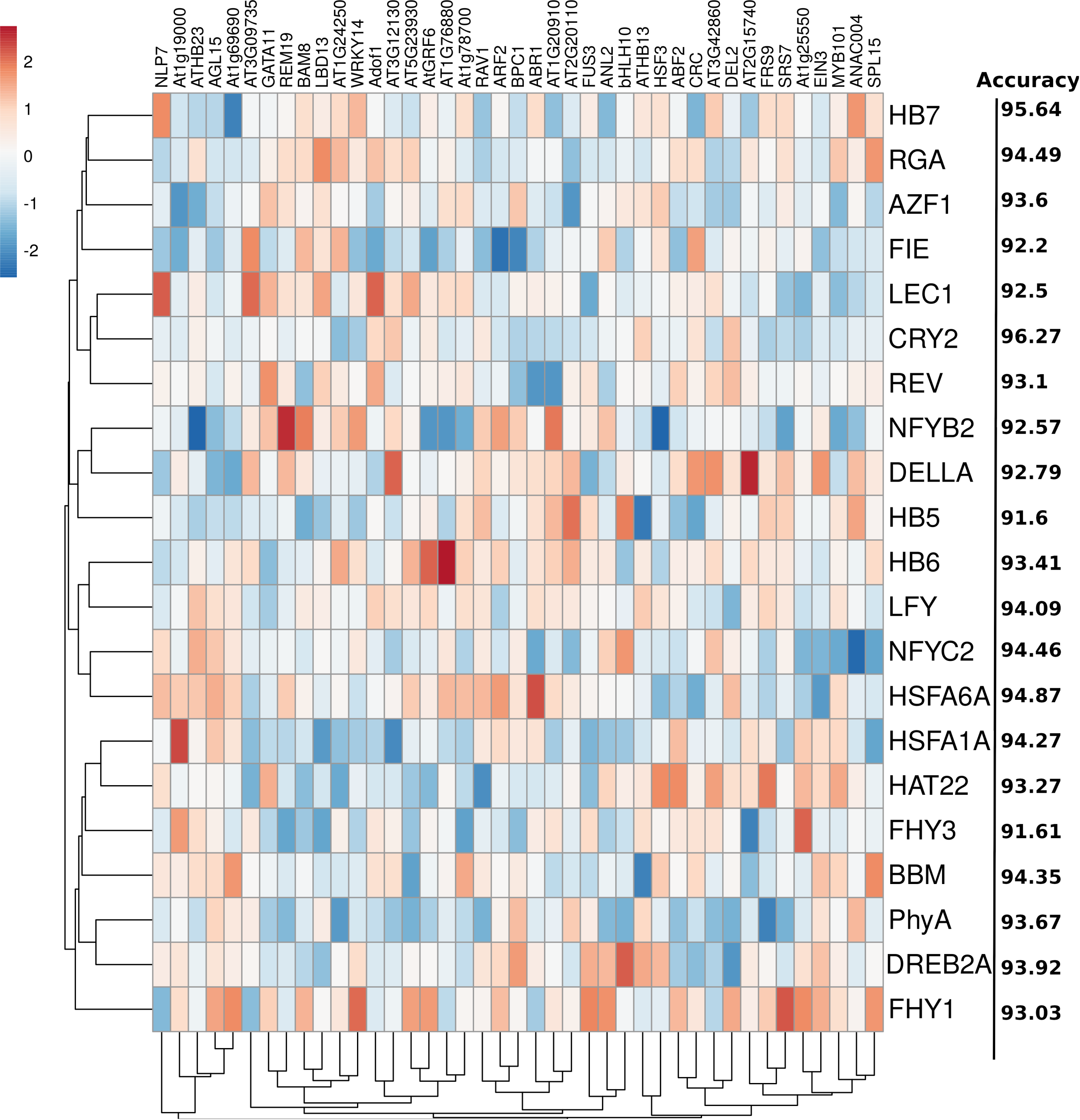
Figure-S3: Structural similarity comparison between representative transcription factors for various TF families from training and testing datasets and their associated accuracy values. This figure illustrates the structural similarity clustering of 41 transcription factors (TFs) used for training and 21 TFs used for testing, along with the accuracy achieved by the PTFSpot algorithm for the testing set TF’s binding regions discovery. The heatmap depicts the structural divergence between the TFs, with red indicating higher dissimilarity and blue indicating higher similarity. The TF FHY1 exhibited the highest structural divergence from all the training data TF members, with an average RMSD of 1.25Å, suggesting a substantial structural difference. Despite this, FHY1 was detected with 93.03% accuracy by PTFSpot, which falls within the claimed performance range and is considered a very good value. In contrast, the PhyA TF showed the highest structural similarity to the training data, with an average RMSD of 0.9Å. Notably, the identification accuracy for PhyA (93.67%) was in a similar range as FHY1. In overall, for all the studied TFs, despite varying structural similarities, the performance range remained the same, clearly indicating that PTFSpot has learned the covariability between structure of the TF and its binding regions rather than simply memorizing the training cases.
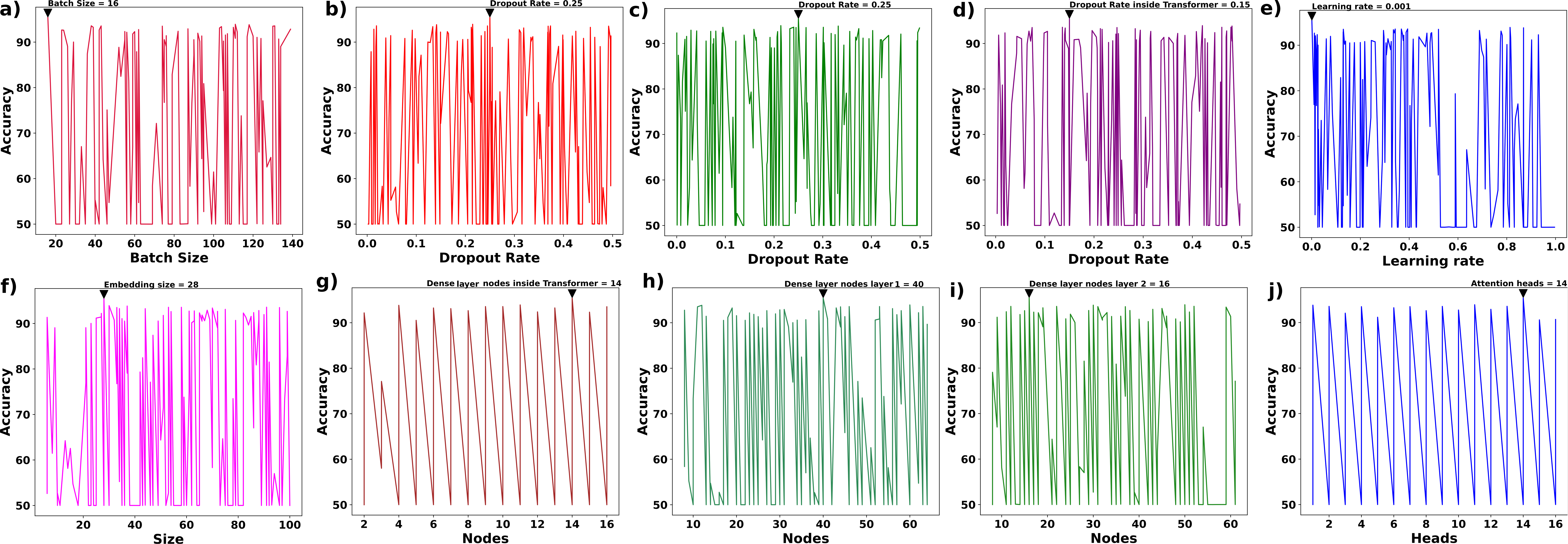
Figure-S4:Optimization results for hyperparamters for transformers model. a) Bacth size optimization, b) Dropout rate optimization inside encoder, c) First Dropout rate optimization, d) Second Dropout rate optimization, e) Learning rate, g) Embedding size, h) Number of units per dense layer inside Transformer, i) Number of units per dense layer 1, j) Number of units per dense layer 2, and k) Number of Attention heads.
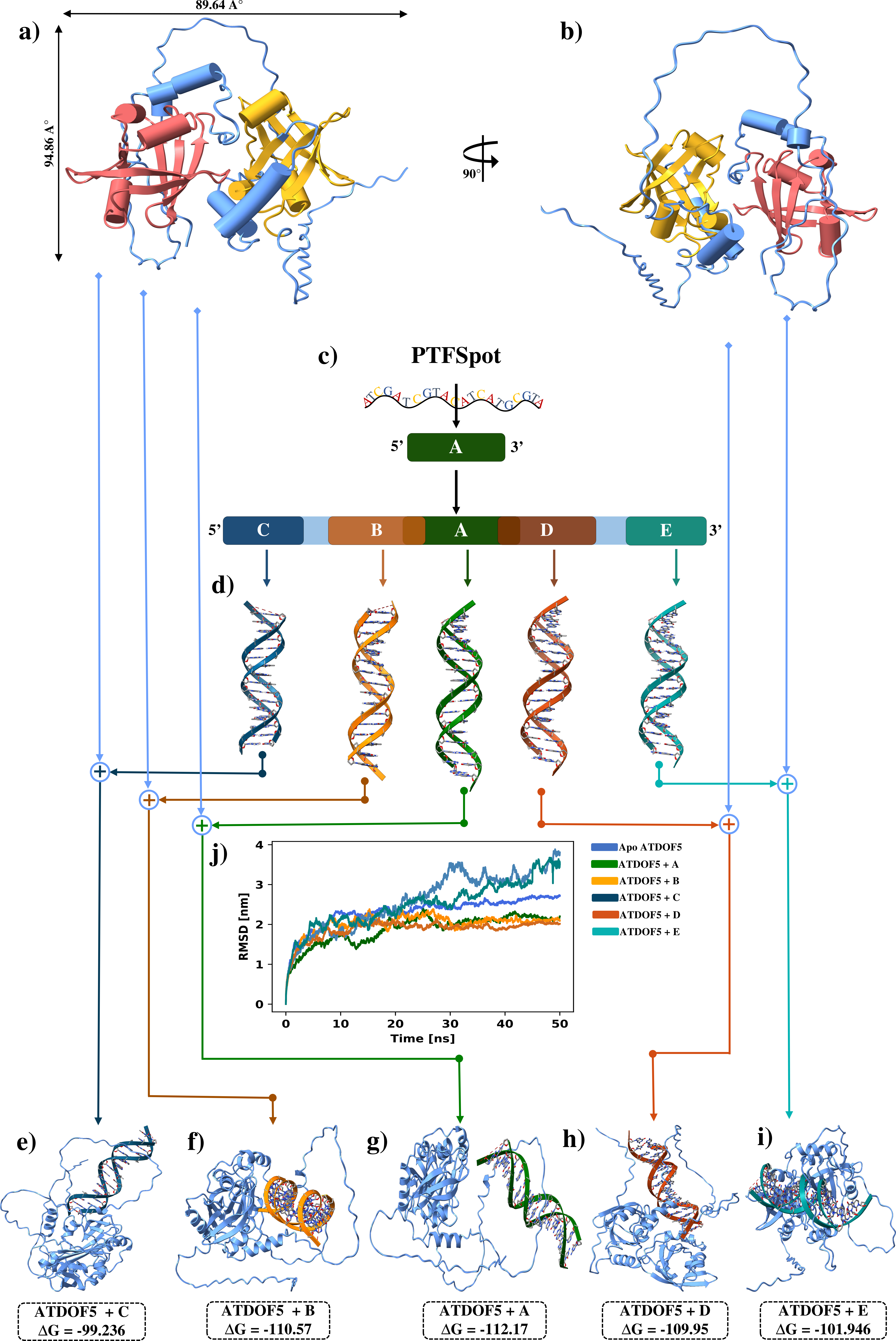
Figure-S5:Molecular docking and simulation analysis between Transcription Factor (TF) and its binding motif. (a) 3D structure of ATDOF5 TF (Arabidopsis thaliana) with two domains (colored in Pale Red and Golden Red). (b) The arrangement of the loops and helices viewed from top after rotation at 90°, (c-d) Prime binding motif identification by PTFSpot for the TF ATDOF5, followed by selection of prime binding motif region as well as regions overlapping with it in its flanks and terminal end regions from both sides (A, B, D, C, and E regions). Respective DNA 3D structures were generated for these regions as the central binding one. (e-i) The docked pose of each tiling-deletion variant DNA with the TF ATDOF5 and their binding affinity. Out of five docked complexes, the prime motif docked complex showed highest binding affinity than other complexes clearly supporting the importance in TF DNA interactions. (j) Molecular dynamics simulation of Apo TF and all five docked complex, the RMSD plot showed that the docked complex of ATDOF5 + region “A” exhibited lowest RMSD value among all, clearly reiterating the above observation that the prime motif region has importance in TF DNA interactions.
Figure-S6:Clustering plot of prime motifs from Dataset “A”.This one covers the prime motifs for 387 transcription factors, and their clustering based on the motifs similarities.
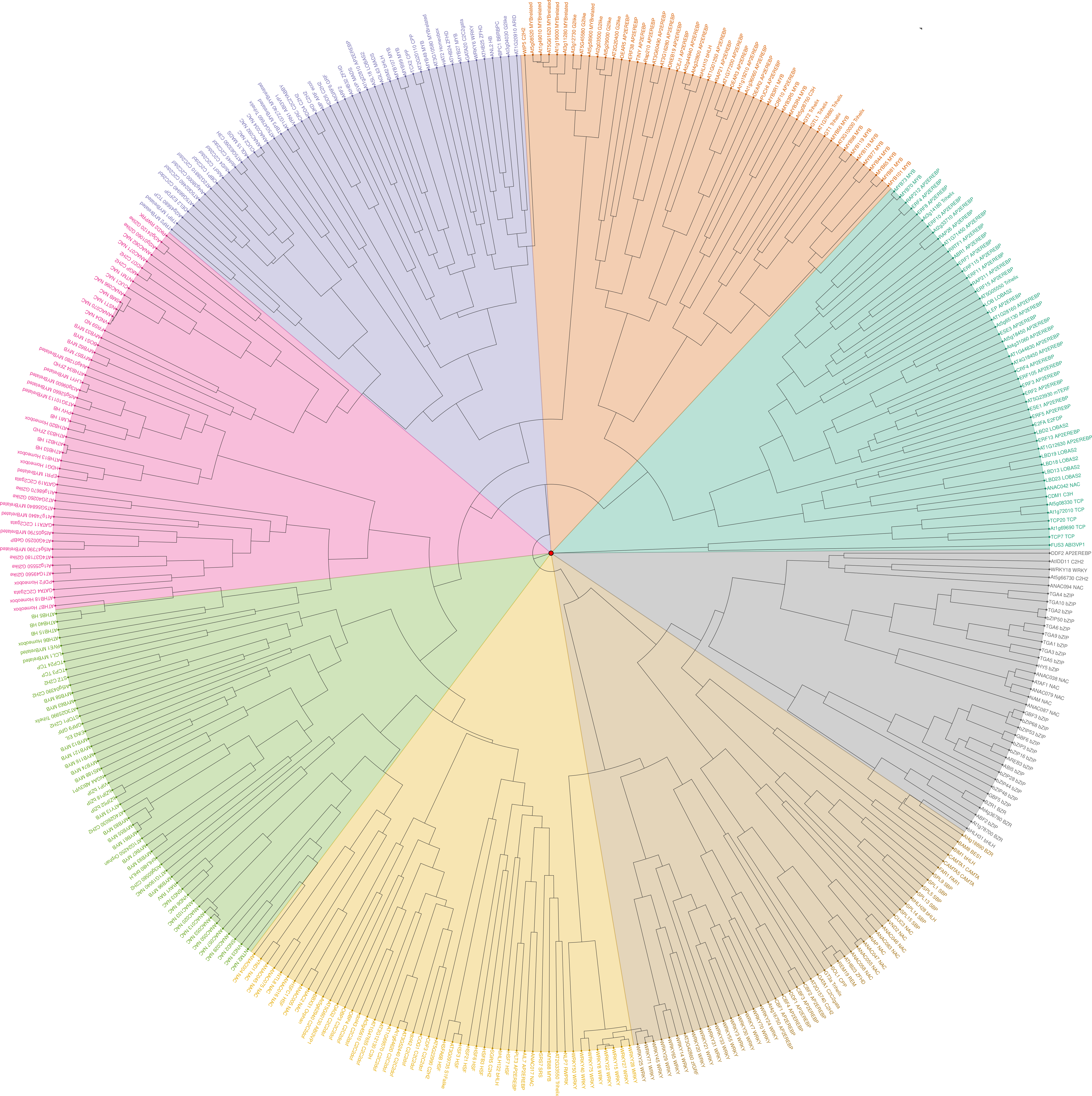
Figure-S7:Clustering plot of prime motifs from Dataset “B”.This one covers the prime motifs for 387 transcription factors, and their clustering based on the motifs similarities.
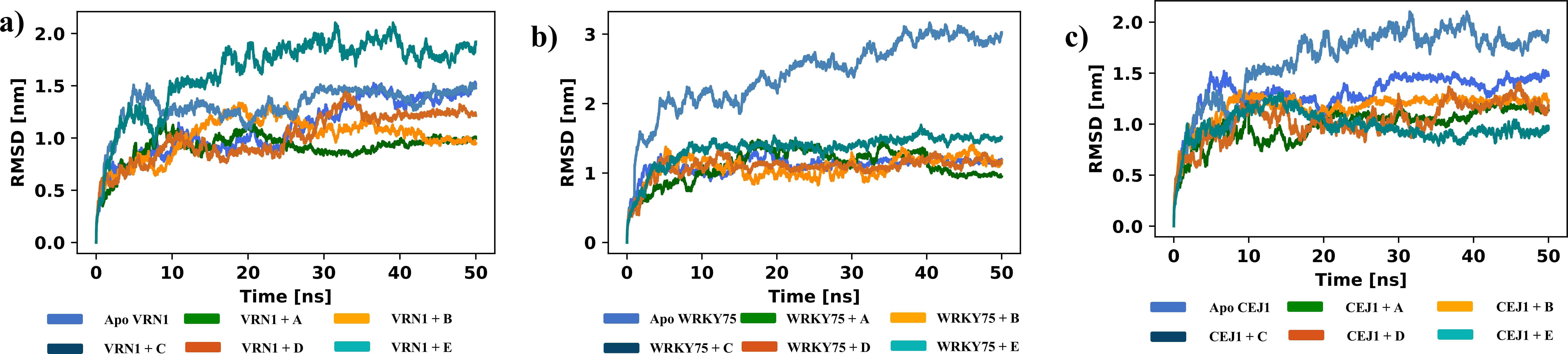
Figure-S9:Molecular simulation analysis between the TFs and their respective binding regions for transcription factors a) VRN1, b) WRKY75, and c) CEJ1.



