Video tutorial of DMRU webserver implementation
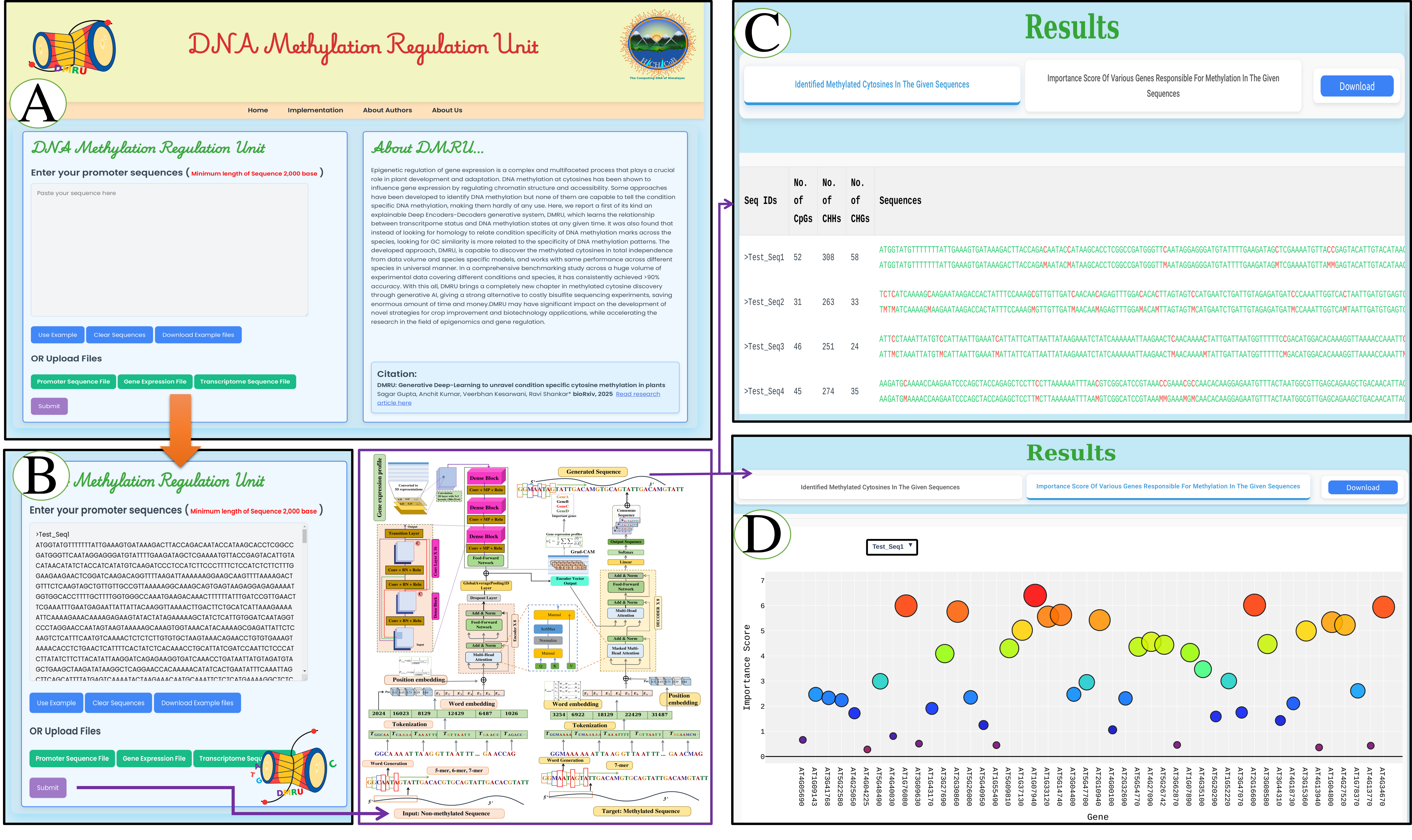
Figure: DMRU webserver implementation. A) Input data box where the user can either paste or load the input files, B) Inputs are DNA sequence in FASTA format and gene expression profile in “tsv” format output from RNA-seq data analysis. C) The methylated cytosines are represented in an interactive form. Download option for the result in the tabular format. D) Also, important feature scoring is implemented and is represented in the form of interactive bubble plot depicting the distribution of scoring distribution for the most influential genes associated with the observed DNA methylation pattern for the user provided target sequence.
Step by Step DMRU webserver implementation
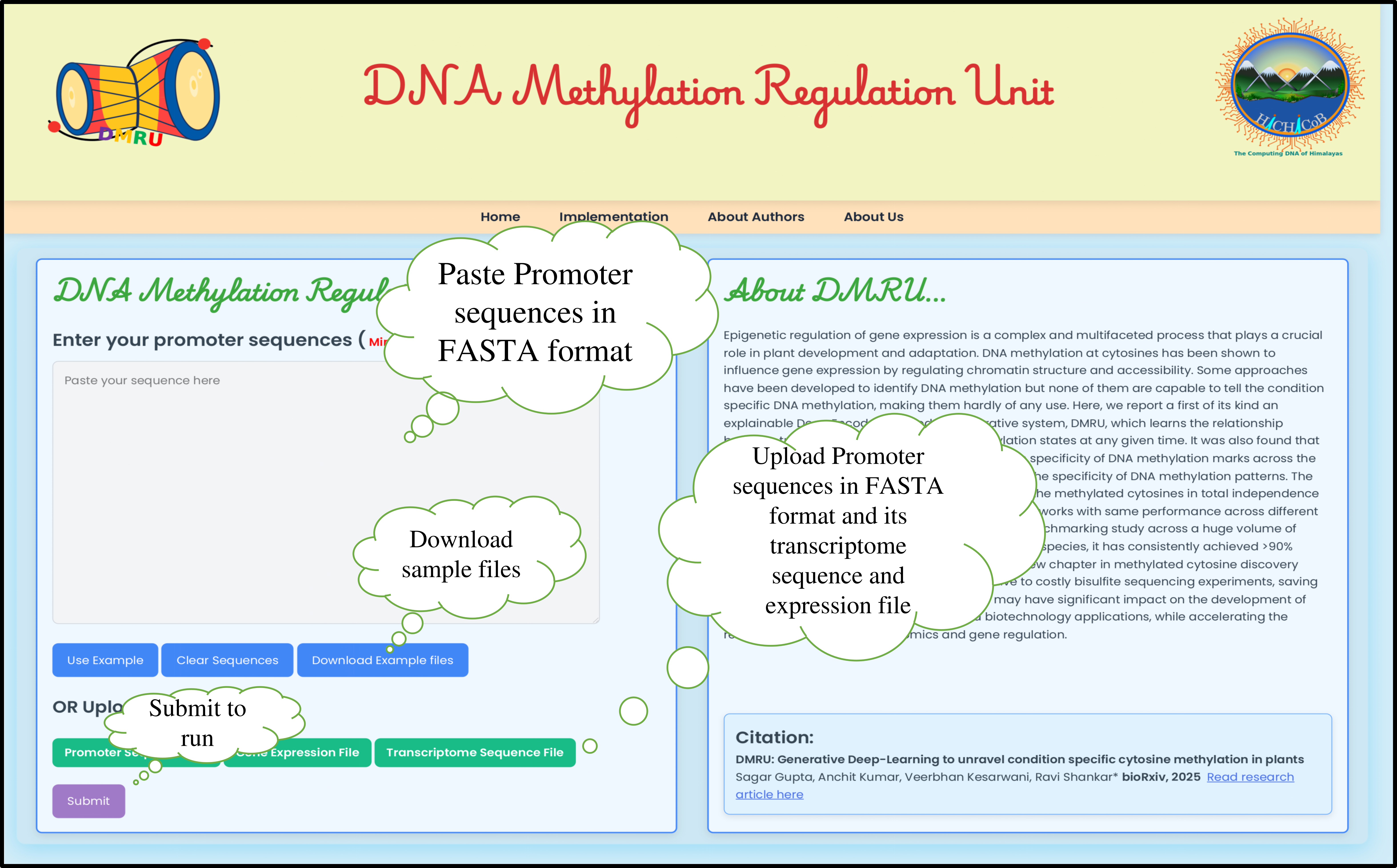
Step1: Input data box where the user can either paste or load the input files. Inputs are DNA sequence in FASTA format and gene expression profile in “tsv” format output from RNA-seq data analysis.
↓
Step2: Upon submission, a loading page will appear until the results are ready.
↓
Step3: The methylated cytosines are represented in an interactive form. Download option for the result in the tabular format.
↓
Step4: Important feature scoring is implemented and is represented in the form of interactive bubble plot depicting the distribution of scoring distribution for the most influential genes associated with the observed DNA methylation pattern for the user provided target sequence.