TAREF (Project funded by DBT, Govt. of India.)
TAREF stands for TArget REFiner. It is a program which refines the microRNA target prediction through incorporation of local interaction information for microRNA and target region in the target mRNA UTR sequence as well as estimates the candidature of being a microRNA target on the basis of intrinsice sequence property around the target site determined by varying dinculeotide density profile. Also by considering dinculeotide density we incorporate the nearest neighbor approach of RNA structure determination methodologies.
Therefore TAREF works with two assumptions:
- Interaction information between microRNA and target is more important than sequence information and such interactions are highly specific to a given microRNA. Experimentally validated interactions can be used to filter out similar interactions in the predicted system to define a preliminary filtering process.
- Varying dinucleotide density profile in the flanking regions around the target region behave as a dynamic and reasonable signature to determine the target sites. Such parameter grossly cover the intrinsic property of the nearby region and sequential arrangement of this varying profile around the target region captures the discrimination.
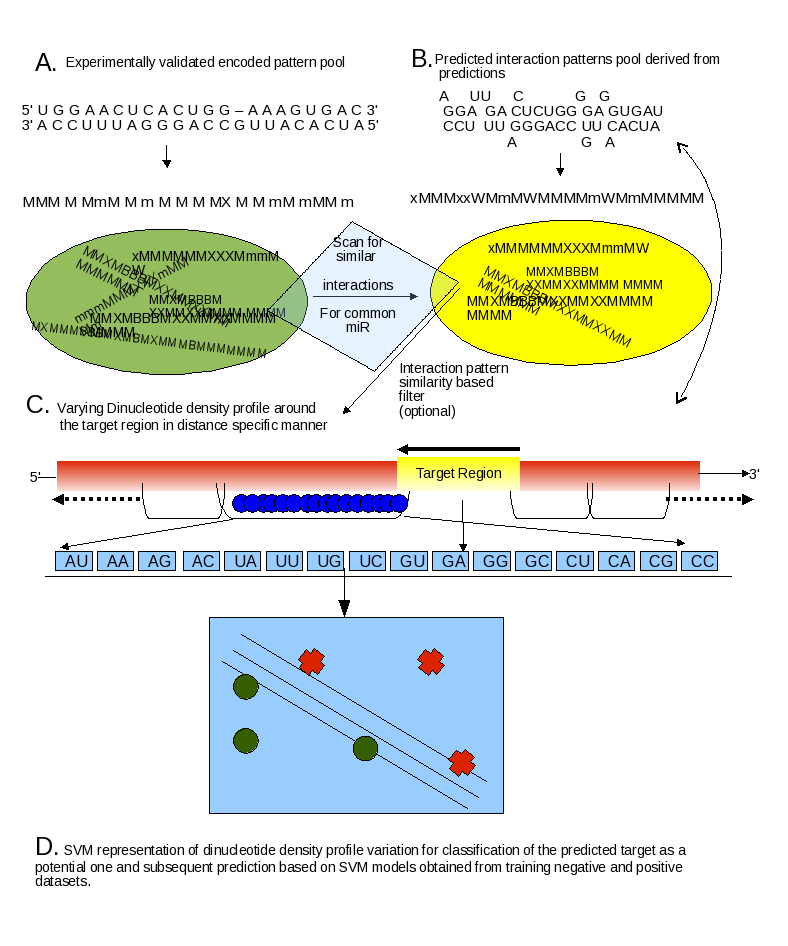
Basically TAREF takes the output of RNAhybrid as the template. TAREF runs in two parts. In first it considers the optional step of filtering through encoded interaction pattern library generated from experimentally reported interactions for human.
TAREF runs in two parts. In first it considers the encoded interaction pattern library generated from experimentally reported interactions for human. These encoded interaction patterns were derived after careful alignment of interaction partners where instead of microRNAs their reverse compliments are taken besides taking a bit extended target region to get more accurate alignment. The parameters used during this step is retained and used during derivation of encoded patterns from the interactions predicted by RNAhybrid. To note here, we absolutely remove the energy cut-off parameters of RNAhybrid as we found several experimental cases which were found at energies much higher than -20 Kcal. This in turn returns lots of output.
Once the encodes are generated from the output of RNAhybrid, the library of experimental encoded patterns is used for scanning for similar interactions in the predicted interactions. At this level itself lots of candidates can be removed. The selected candidates are subjected to our support vector scan. This support vector machine utilizes the information of intrinsic sequence property of the flanking regions around the target region in the form of varying dinculeotide density profile at various intervals. We found that the peak discrimination capacity of SVM models was observed when intervals of 20 bases were considered.
Our methodology on which this software server is based, has attained accuracy level of ~90% with high level of sensitivities and specificities and Area Under Curve for ROC analysis were above 0.9. Details are present in performance tab.