|
|
Home Research Software & Web server Lab Members Publications Workshop/Conference GitHub Careers
|
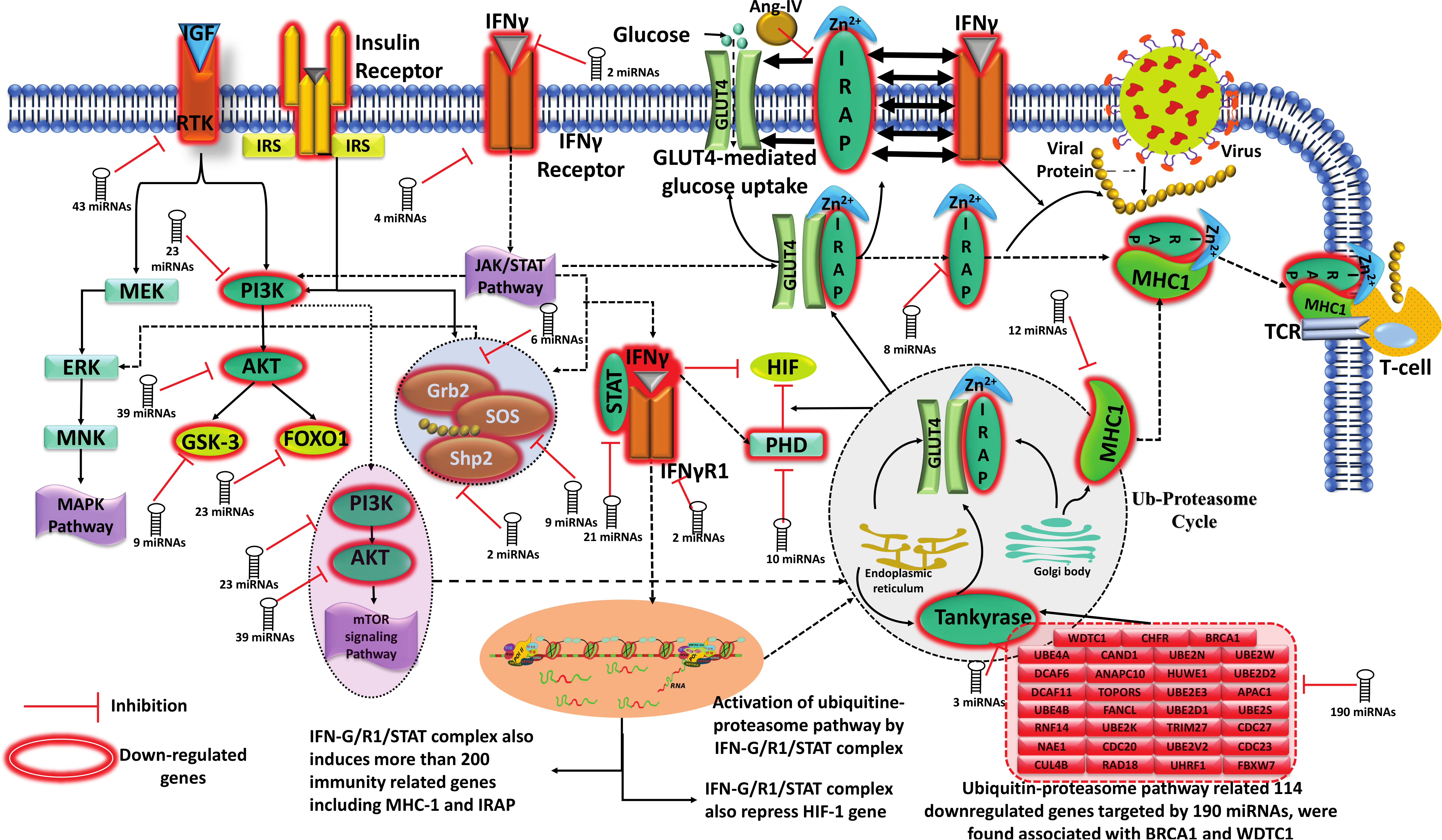
Research highlighted in News/Media Blog @SCBBResearch HighlightsRBP-MiRNA interactions that reveal post-transcriptional regulation of gene expression and also reveal their role in the SARs-COV2 host miRNAomeDROSHA / DICER are common players in the formation of miRNA through the miRNA biogenesis process. In addition, several other factors are also actively involved in the biogenesis of miRNA. Comprehensive research such as association between RBP and their network with miRNA and Gene expression analysis of CLIPseq and RNAseq high-throughput data shows that different RBP combinations are actively involved in miRNA gene expression regulation. figure-1: miRNAs regulate several genes critically involved in the cross talk of pathways related to IFN-Gamma signaling, Insulin/IGF/ATK/mTOR signaling, and associated Ubiquitin-Proteasomal pathways in SARS-CoV-2 patients. IFN-Gamma appears very important molecule here. It regulates Insulin regulated aminopeptidase gene, IRAP. IRAP is critical for glucose metabolism as it induces translocation of GLUT-4. IRAP in influence of IFN-Gamma performs its peptidase function to generate antigens from viral peptides and associates with MHC Class-I to present the peptide to CD-8 T-cells, causing immune response. Besides this, IRAP is also blocked by Angiotensin IV which was found elevated in Covid19 infection. IFN-Gamma system also interacts with IGF/RTK/IR/PI3K systems whose impact is wide, influencing MAPK pathway as well as Insulin/Glucose signaling through PI3K/AKT/mTOR signalling pathways. PI3K/AKT system is also helps in induction and protein targeting for IRAP-GLUT-4 vesciles from ER/Golgi complex through Ubiquitinylation/deubiquitinylation cycles. Tankyrase is a critical protein here which directly interacts with IRAP/GLUT-4 to translocate them. Tankyrase itself is controlled by a series of genes involved in Ubiquitinylation-proteasomal pathways. The same system also regulates production of MHC Class-I and associated immune response. IFN-Gamma system is also involved here with JAK/STAT system where it forms complex with its receptor R1 and STAT and represses the transcription of HIF gene as well as enhances the PHD gene which degrades HIF-alpha and restores normal oxygen conditions. This IFN-Gamma/R1/STAT complex also induces transcription of critical genes involved in immunity, and induces MHC-I and IRAP expression. Very interestingly, most of these critical genes were found downregulated by number of identified miRNAs, starting form IFN-Gamma, its receptor R1, RTK, PI3K, AKT, IRAP, MHC-I, Tankyrase, PHD, and 114 critical genes of Ubiquitin-Proteasomal pathways, capturing the observed pathophysiological signs of Covid19 infection. This axis emerges as a very promising one for therapeutic interventions against SARS-CoV-2. Beside this miRNA sequences also target Insulin/IGF/P3K/AKT, and Ub-proteasome pathways and regulate survival of infected cell through downregulation via crosstalk to eachother. This novel discovery may be a promising approach toward therapeutic discovery against SARS-nCOV infection. |
||||||||||||||
| Developed by Heikham Russiachand Singh (past member). Maintained by Ganesh Panzade SCBB, Biotech Division. | |||||||||||||||
|
Copyright © 2016, Institute of Himalayan Bioresource Technology.
|
|||||||||||||||