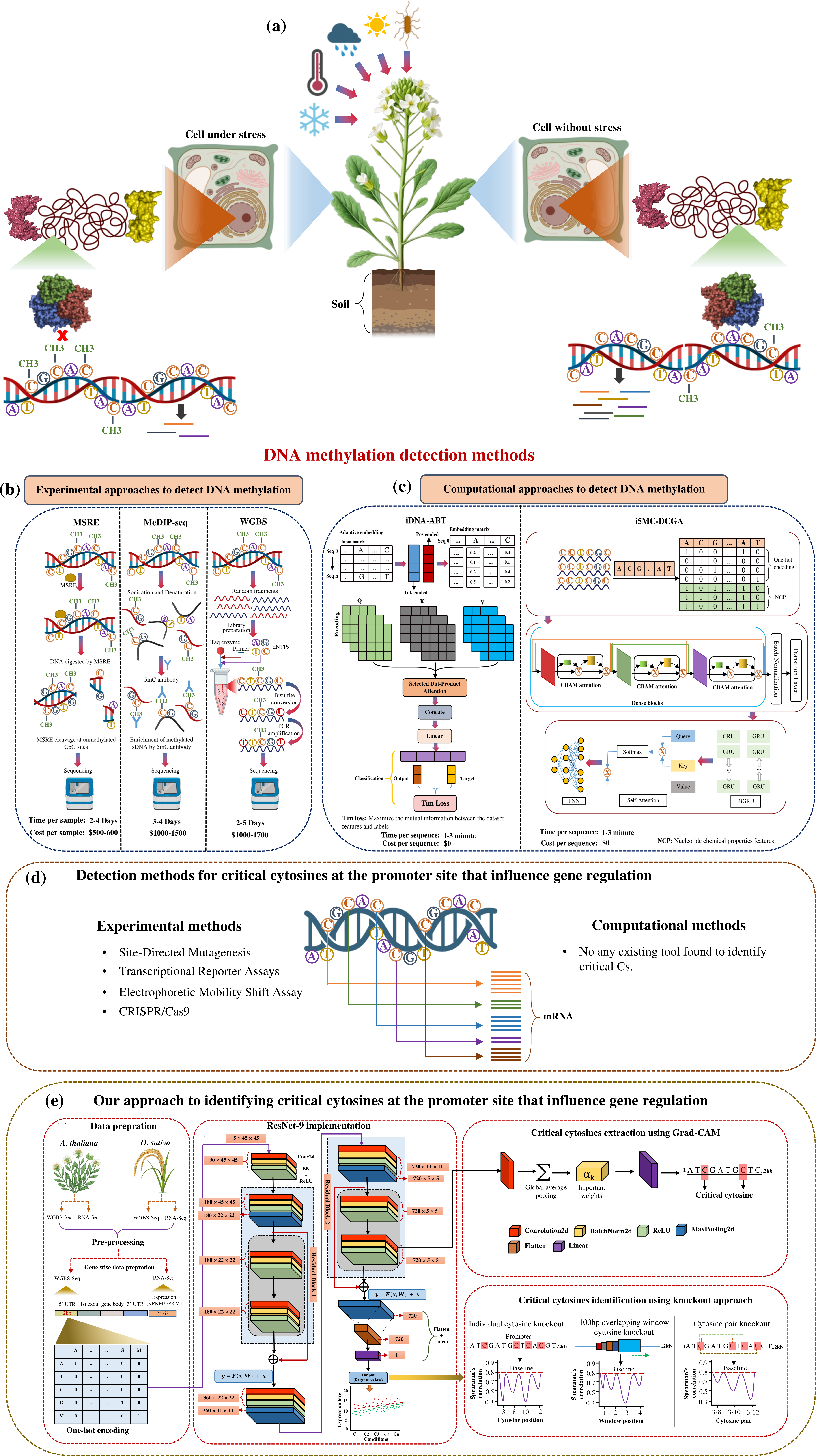
Overview of the study
(a) Stress-dependent regulation of gene expression by DNA methylation; Left: Under normal conditions, unmethylated DNA permits transcription factor (TF) binding, enabling gene expression, Right: Under biotic/abiotic stress, DNA methylation (red circles) inhibits TF binding, suppressing gene expression. (b) Experimental methods for DNA methylation detection. (c) Computational approaches for methylation detection. (d) Limitations in studying single-cytosine effects, Current experimental methods can assess individual cytosine impacts but are costly, time-consuming, and condition-specific. No computational tools yet exist to predict expression-level effects of specific cytosines. (e) Our solution: Critical cytosine analysis, pipeline for identifying and validating individual cytosines that directly influence gene regulation, bridging the gap between methylation status and expression outcomes.
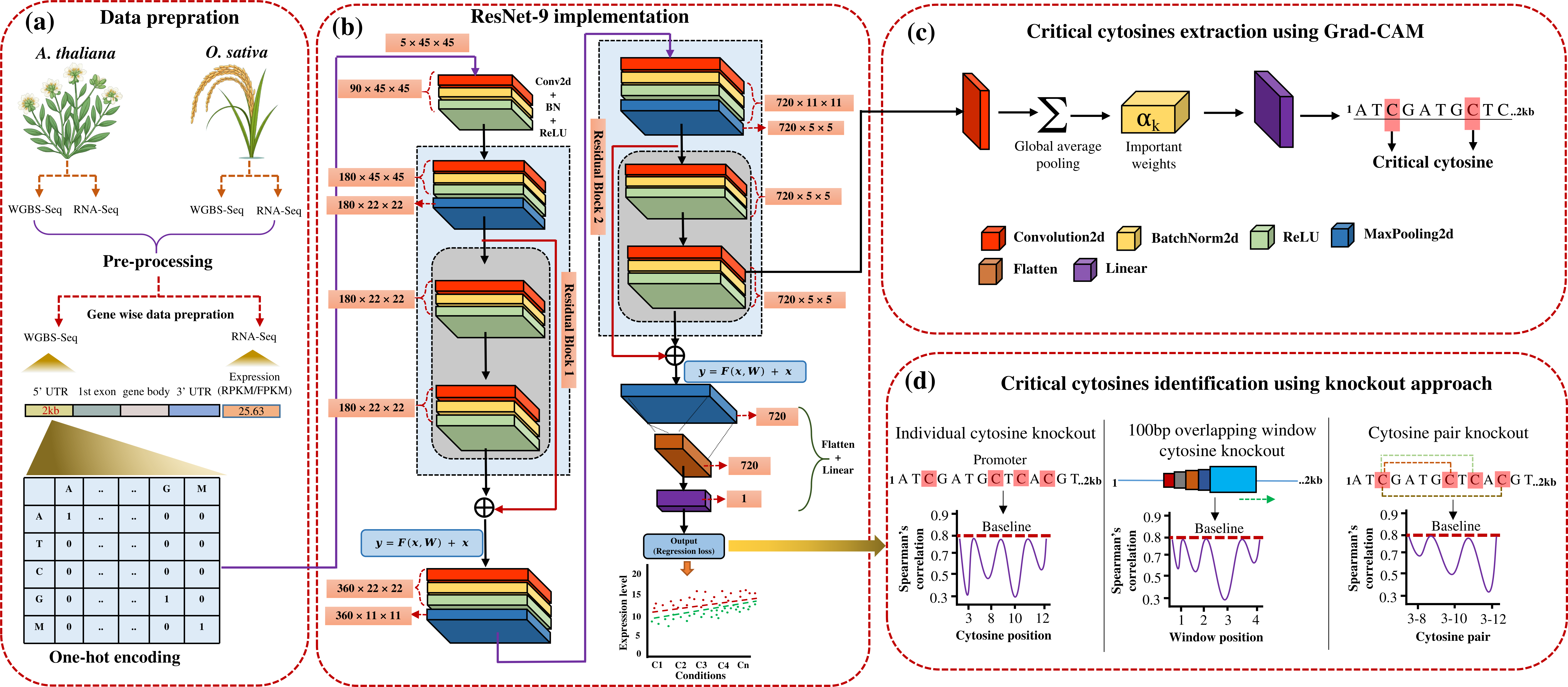
A deep learning-based framework for expression prediction and critical cytosine identification using Grad-CAM and knockout approach.
(a) The data preparation workflow includes WGBS-Seq and RNA-Seq datasets from A. thaliana and O. sativa under multiple conditions. For each experimental condition, gene-specific promoter sequences (2kb upstream) are processed by extracting methylation patterns and corresponding gene expression (FPKM/RPKM) levels, which are subsequently transformed into one-hot encoded representations. (b) Implementation of the ResNet-9 deep learning model. The architecture processes 5x45x45 input tensors representing the one-hot encoded promoter sequences. Residual blocks enhance feature learning, and the final output is a regression score representing predicted expression. (c) Critical cytosines are extracted using Grad-CAM. Feature importance weights are computed and mapped back to the input promoter sequence to identify cytosines that significantly contribute to gene expression prediction. This integrated approach enables systematic identification of functional cis-regulatory elements across species and conditions. (d) Critical cytosines are validated using a knockout analysis. Three strategies are illustrated: individual cytosine knockout, 100 bp overlapping window knockout, and cytosine pair knockout. The effect of each knockout on model performance is quantified by changes in Spearman's correlation with baseline predictions.