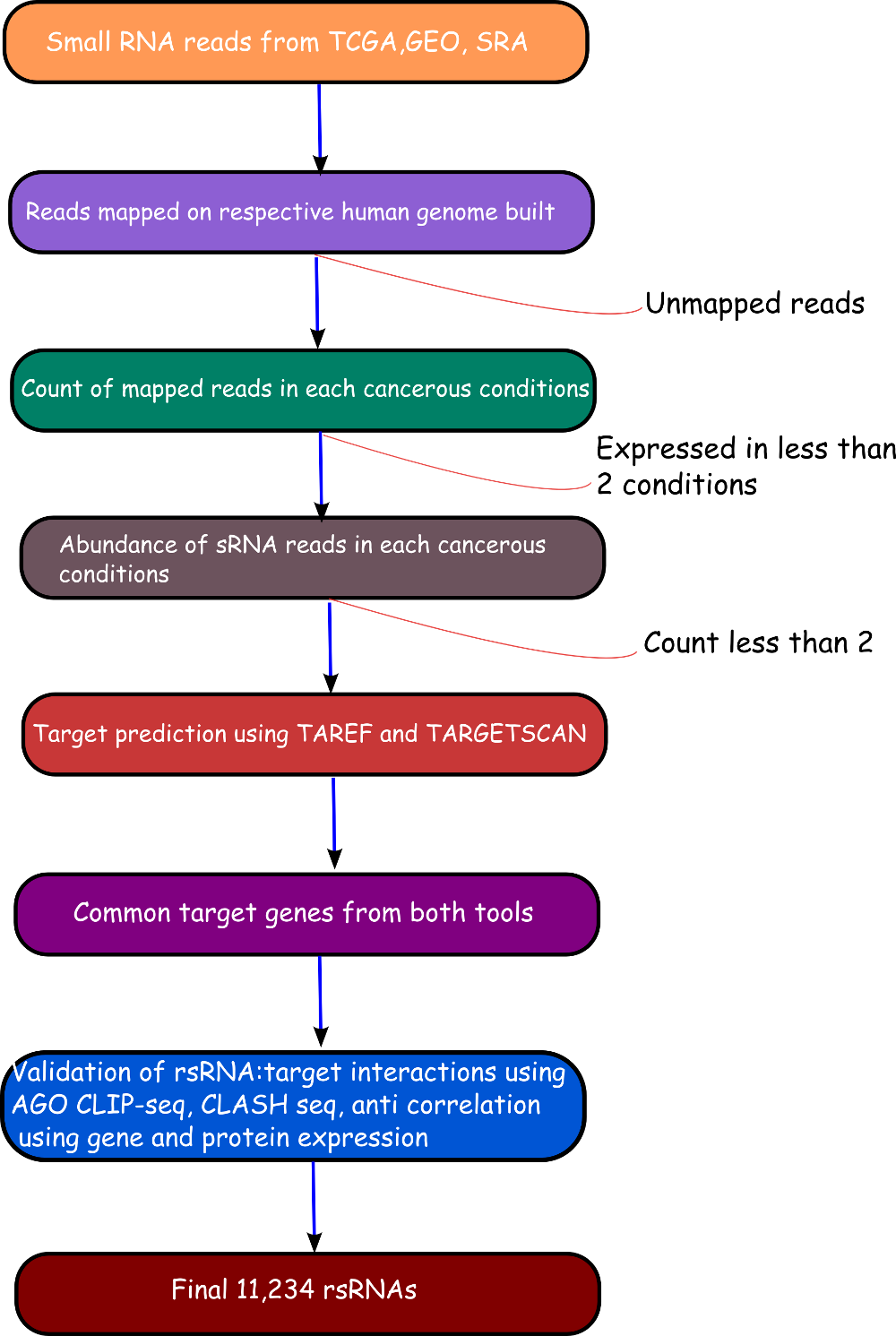
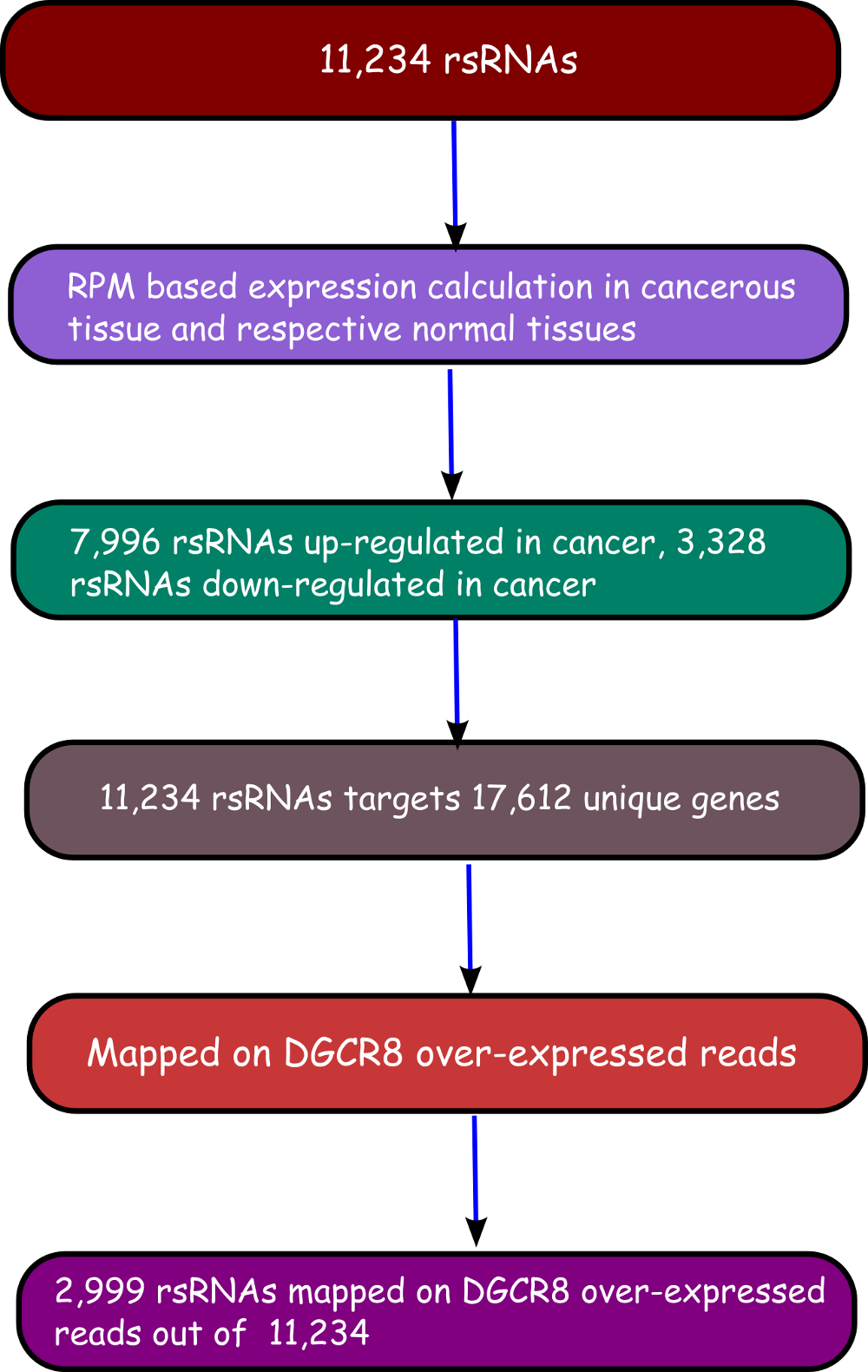
We have performed a comprehensive study and identified 11,234 novel small regulatory RNAs (rsRNAs) which regulates about 17,000 unique genes. The role of these 11,234 novel rsRNAs were studied with respect to 25 different cancerous conditions. Data for these cancerous conditions was downloaded from The Cancer Genome Atlus (TCGA) and Gene Expression Omnibus (GEO) . Data from 4,997 individuals were considered for the analysis which results in 260 Gb of processed data. Total raw data for the complete analysis was about 20Tb.
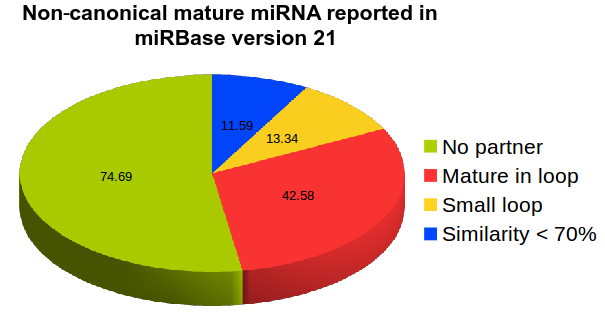
It was believed that miRNA/sRNA biogenesis depends on presence of terminal loop, where mature miRNAs arises from stem region when Dicer cuts the double stranded stem region of stem loop structure with two overhangs. After introduction of small RNA based NGS-sequencing technologies many new miRNAs were reported in miRBase which do not follow the above criteria such as presence of terminal loop region (hsa-miR-181d, hsa-miR-141) absence of pairing mature miRNAs (in about 45% of total mature miRNAs reported (hsa-miR-944, hsa-miR-378d-2), mature miRNAs coming from loop regions instead of stem region (hsa-miR-451a, hsa-miR-7111), rsRNA reads mapping on other region instead of reported known miRNAs (hsa-miR-5680, hsa-miR-5697), mature miRNAs with no overhangs (hsa-miR-7108). These miRNAs suggest that following only the canonical or traditional pathway for miRNA biogenesis has become a bottleneck in identification of miRNAs creating bias towards analysis, and report only those miRNAs which follows the same patterns of already reported miRNAs in miRBase. Small RNAs have been reported as an important regulatory component of genome and considering only those rsRNAs as miRNAs which follows old traditional biogenesis patterns narrows the understanding of molecular mechanisms of cell growth and development.

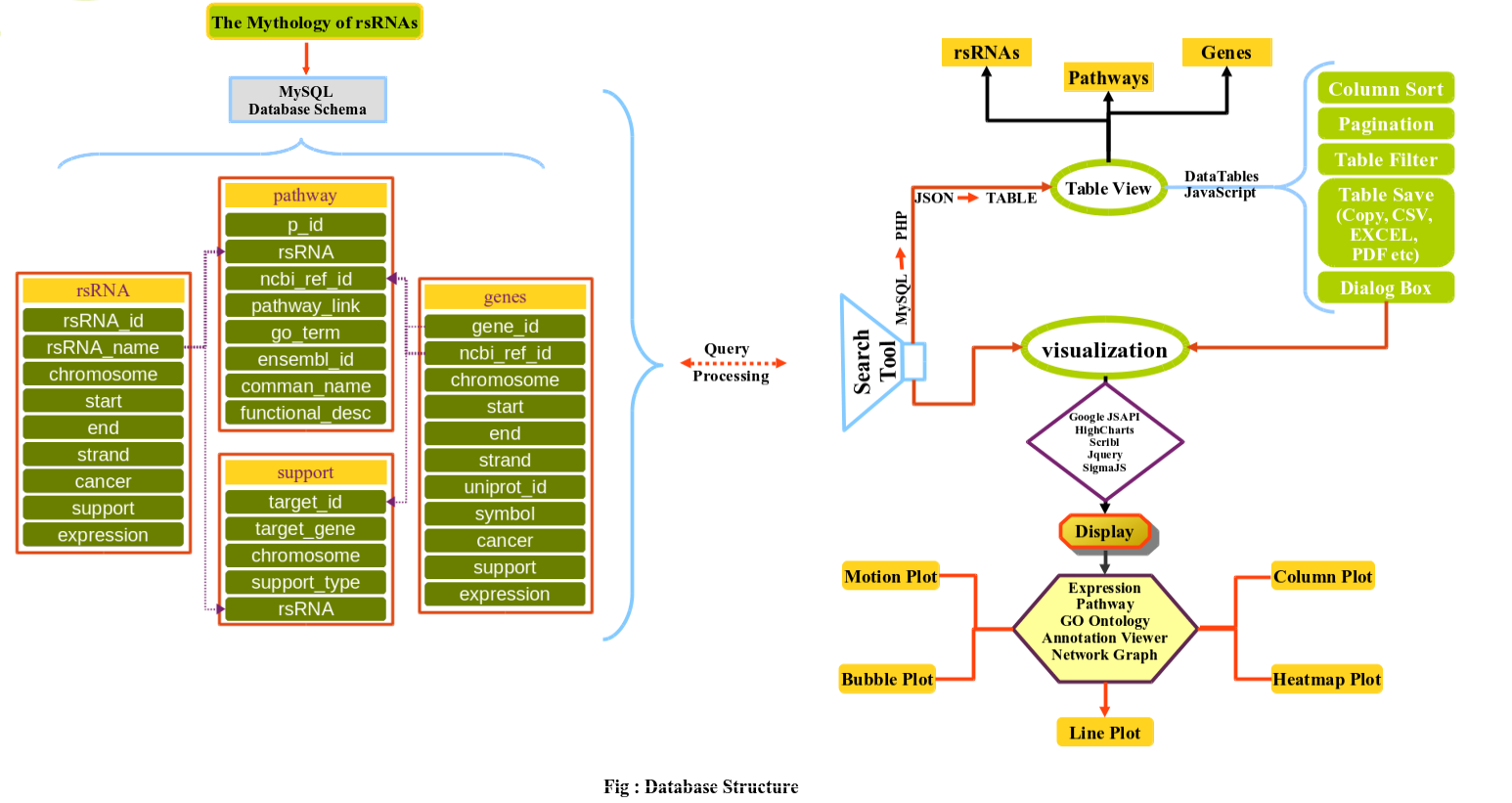
Methodology

Results obtained so far

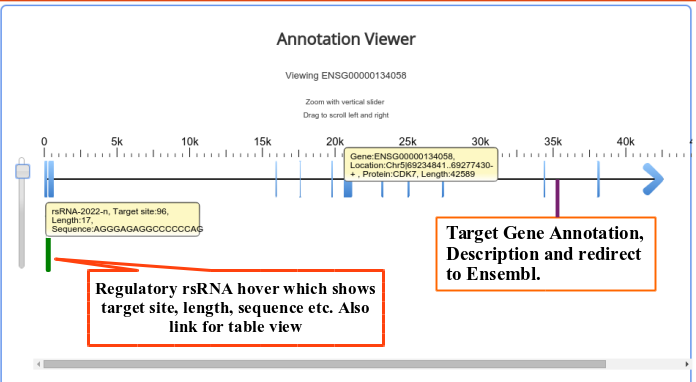
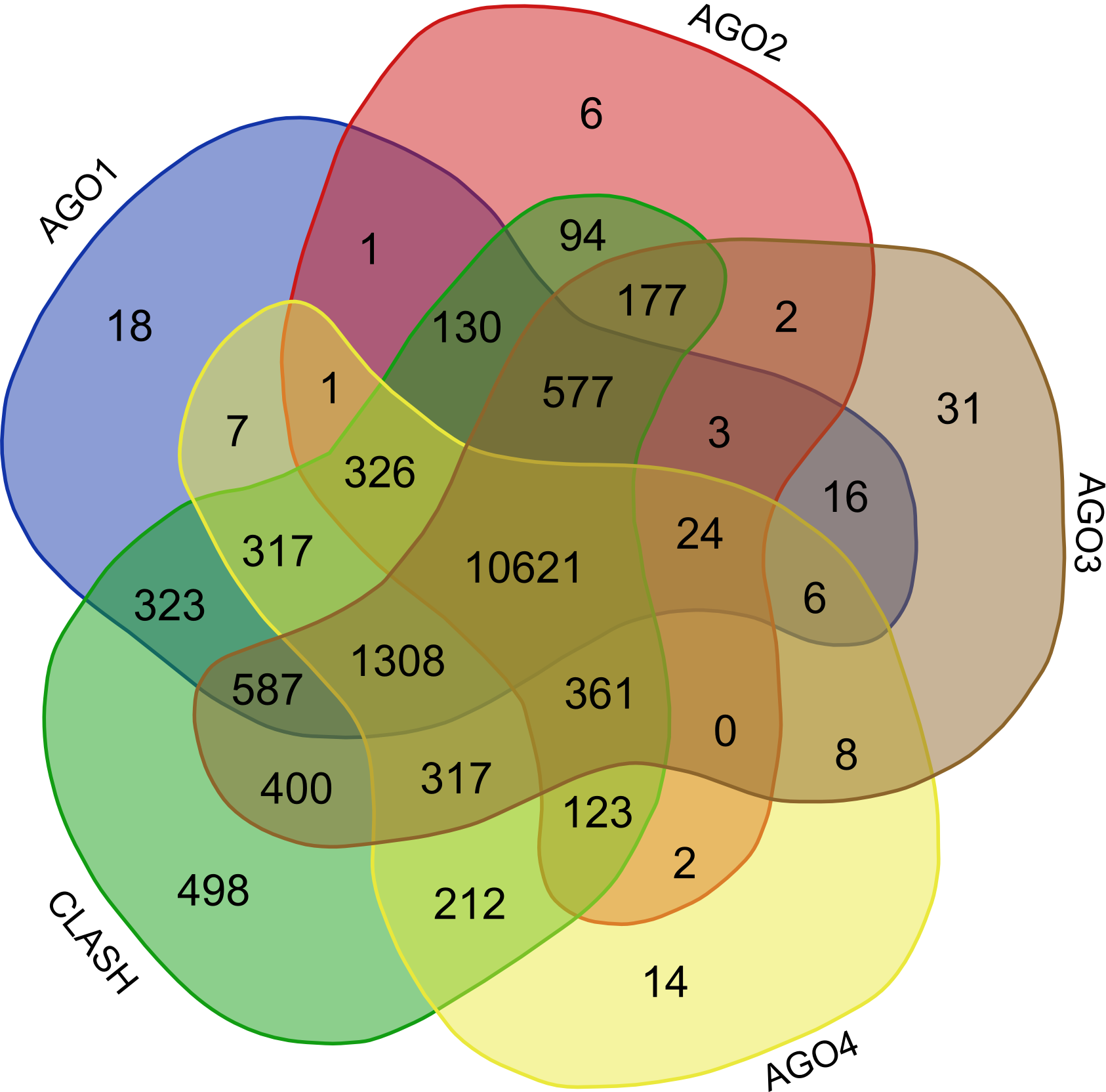
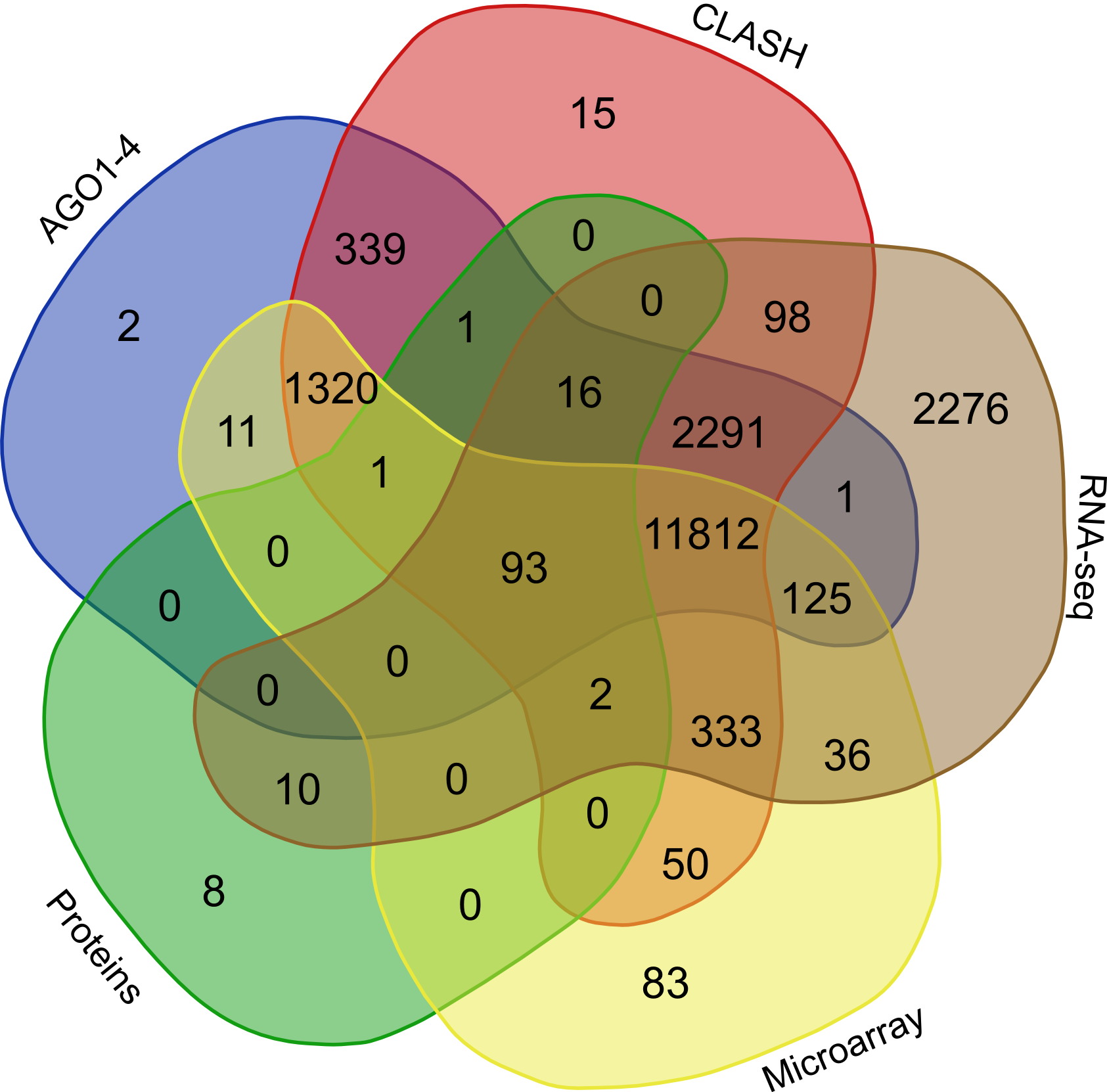
Validation of rsRNA:target inetaractionswas done by searching the inetractions in the Argonaute sequencing data downloaded from starBase version 2. A total of 149,344, 150,049, 145,561 and 148,251 novel putative small regulatory RNA:target interactions were identified for 8,036, 6,247, 8,342 and 7,196 unique rsRNAs and 14,265, 12,448, 14,438 and 13,674 unique targeted genes, for AGO1, AGO2, AGO3 and, AGO4-CLIP sequencing data, respectively.
CLASH-seq data was also used for the identification of rsRNA: target interactions. As compared to CLIP-seq, CLASH-seq data gives more precise information about the interactions as the interactions are precisely arrested through ligation of the target and targeting small RNA. Using CLASH-seq data, 16,371 unique genes were found being targeted by 10,048 putative rsRNAs, comprising 474,770 unique target interactions.

Furter to validate targets, anti-correlation based validation study was performed using sRNA expression (RPM) and expression of target genes (RPKM) obtained from TCGA and GEO. A total of 3,013 cancerous and normal tissue based experimental conditions were considered. For those cancerous conditon in which RPKM data was not available, microarray absed expression date from TCGA (for 137 patients and normal conditions) were used. A high level of agreement was observed between different methods of validation for the identified rsRNA:target interactions.

These novel sRNAs were found regulating many genes including some important genes involved in cacer like BRCA2, p53, Rb, Myc, 14-3-3 epsilon, CycD and CycE.

From the mapped data, it was found that these small regulatory RNAs have multiple biogenesis loci mainly belonging to repetitive elements (48.22%), Intronic region (36.02%) and ncRNA (9.31%) regions. Many of the known miRNAs have been reported from intronic region, in-fact 46.03% (866 out of 1881) miRNAs, in current version of miRBase are from the intronic regions.

Several rsRNAs were found originating from the Alu elements. While observing the distribution profile of these sRNAs across the length of Alu consensus, it was observed that, these rsRNAs follow a conserved pattern of biogenesis for a number of different experimental conditions. A video compilation of Alu derived sRNA expression profile variations for the different cancer states and normal tissues has been made available to showcase this behavior of Alu derived sRNAs .
These identified rsRNAs were clustered using four different methods namely 1) seed region similarity, 2) sRNA length and Argonatue association, 3) all smaller sRNAs are covered by a single long sRNA and, 4) Expression based clustering. These clustering was done to identify rsRNAs sharing similar properties. rsRNAs in a cluster were found targeting same/similar gene, regulating similar pathways and similar functions. Expression based clustering results in 362 distinct clusters which includes 7,292 rsRNAs these rsRNAs share high co-expression with each other ("r" > 0.5) calculated for 4,997 experimental conditions.
For clusters identified form a given approach overlapping clusters were identifed. High commonalities between the clusters generated based upon different properties suggest a degree of similarity and relationship between the two properties. This way a 4X4 matrix was generated for similarity scoring between the clusters of above mentioned four types. It was found that expression based clusters shared ~82% similarity with the clusters generated using coordinates bound based clustering, 22% with seed clusters and 0.23% with length based clustering. Coordinates bound based clusters displayed 77% overlap with seed based cluster and 0.09% clusters overlap with length based clustering. This analysis suggested that the three clustering methods based on seed region, coordinates bound and expression similarity agreed with each other, where the co-expressed sRNAs were also similar to each other in terms of genomic coordinates, which also shared a good amount of common targets.

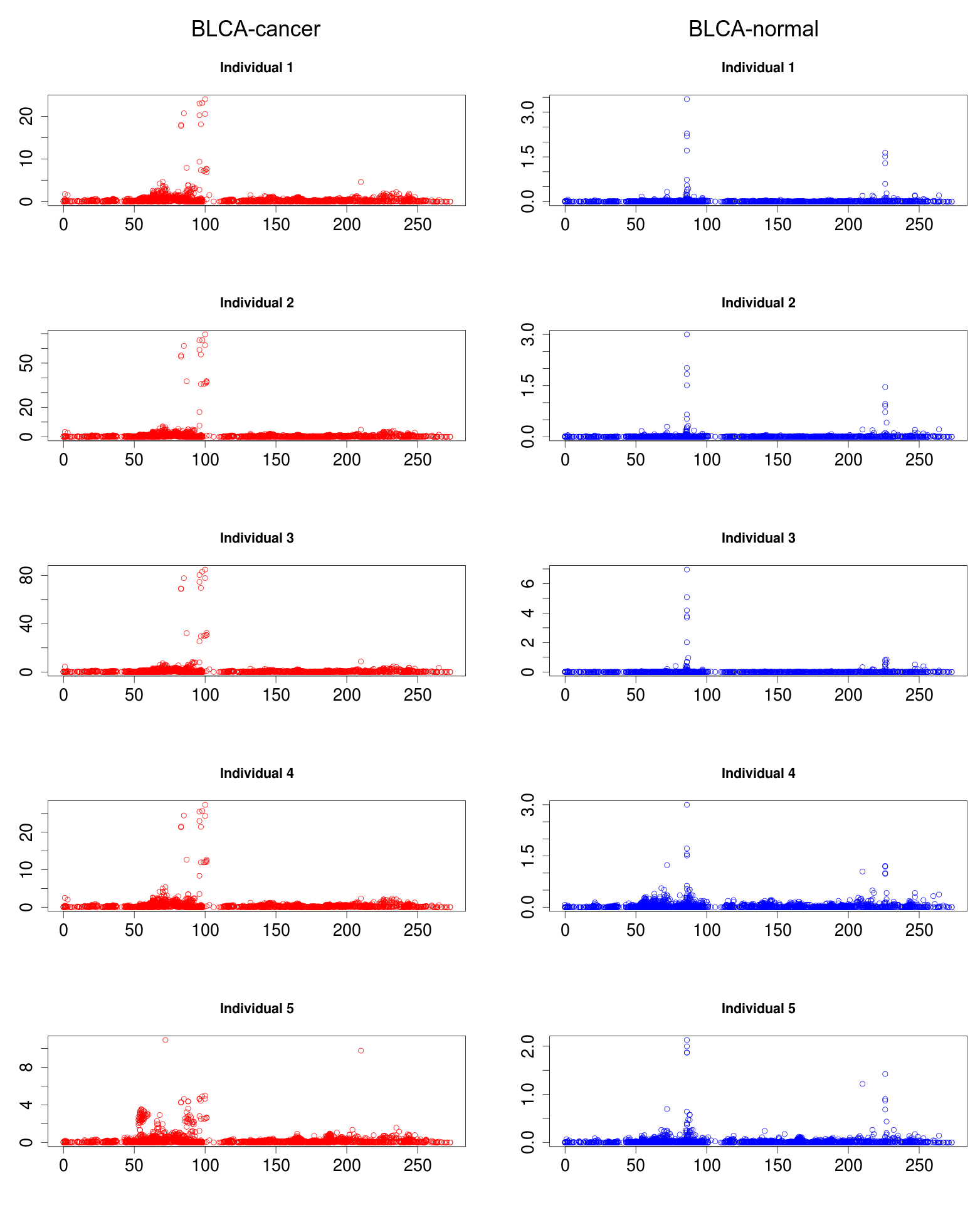
Several novel regulatory small RNAs were found significantly differentially expressed between in cancerous conditions and in a normal condition. The differential expression was evaluated across large number of individual samples, followed by t-test for significance between normal sample sets and cancer patients sample sets, for every cancer condition.
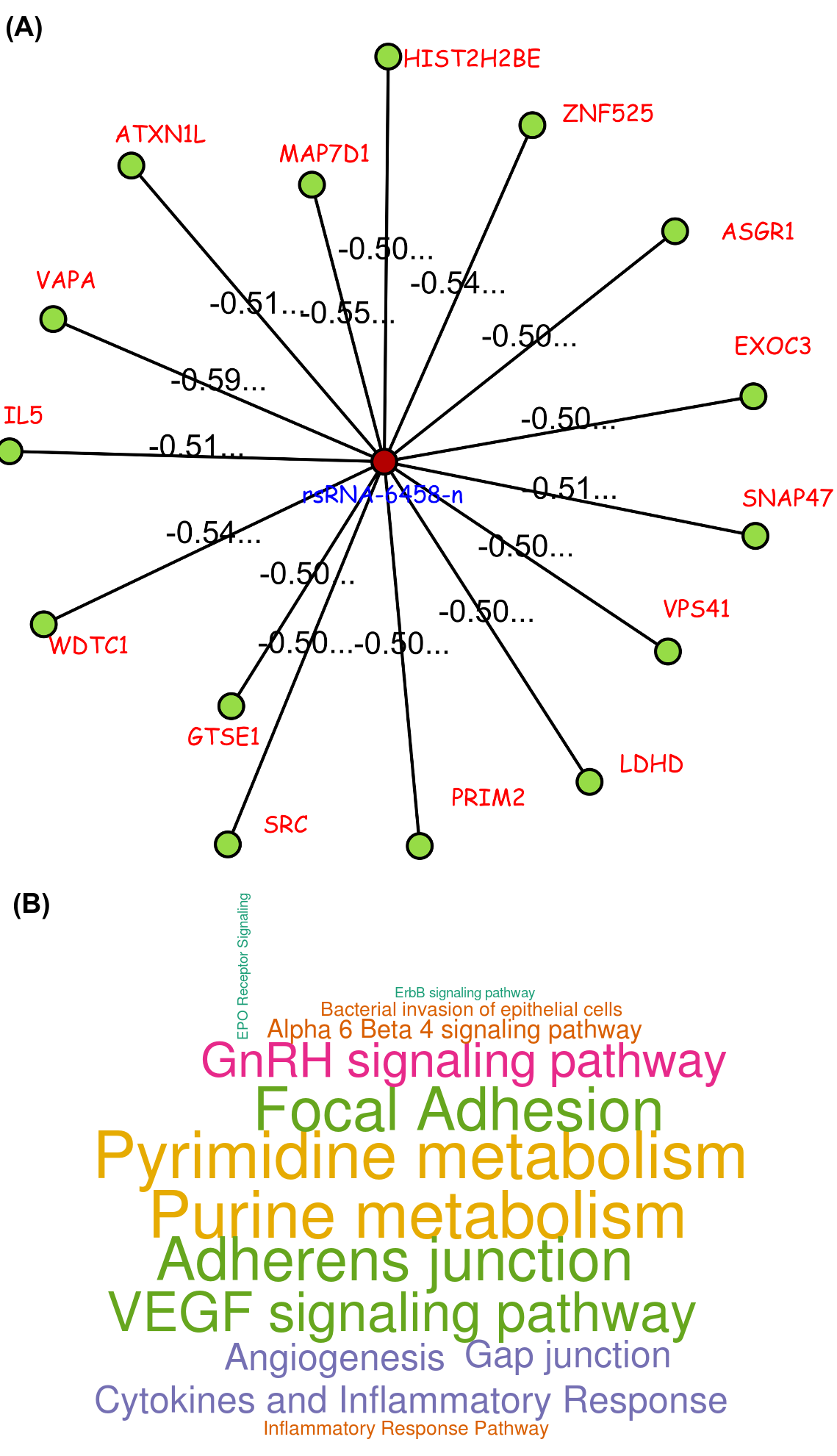
A small regulatory RNA 9881 . This small regulatory RNA was found overexpressed in almost all cancer states studied here, suggesting about some central points being affected by this regulatory small RNA. There were 20 different target genes which were found strongly negatively correlated to its expression. A closer analysis revealed that the target genes were enriched for pathways critical for cell development and cancer, at the interfaces of diverse pathways (apoptosis, cell death, p53 signaling, hiv-1 nef, caspase cascade, TLR, TNFR-1 signaling and FAS pathway), reasoning why the regulatory small RNA 9881 was found abundant in most of the studied cancer conditions.
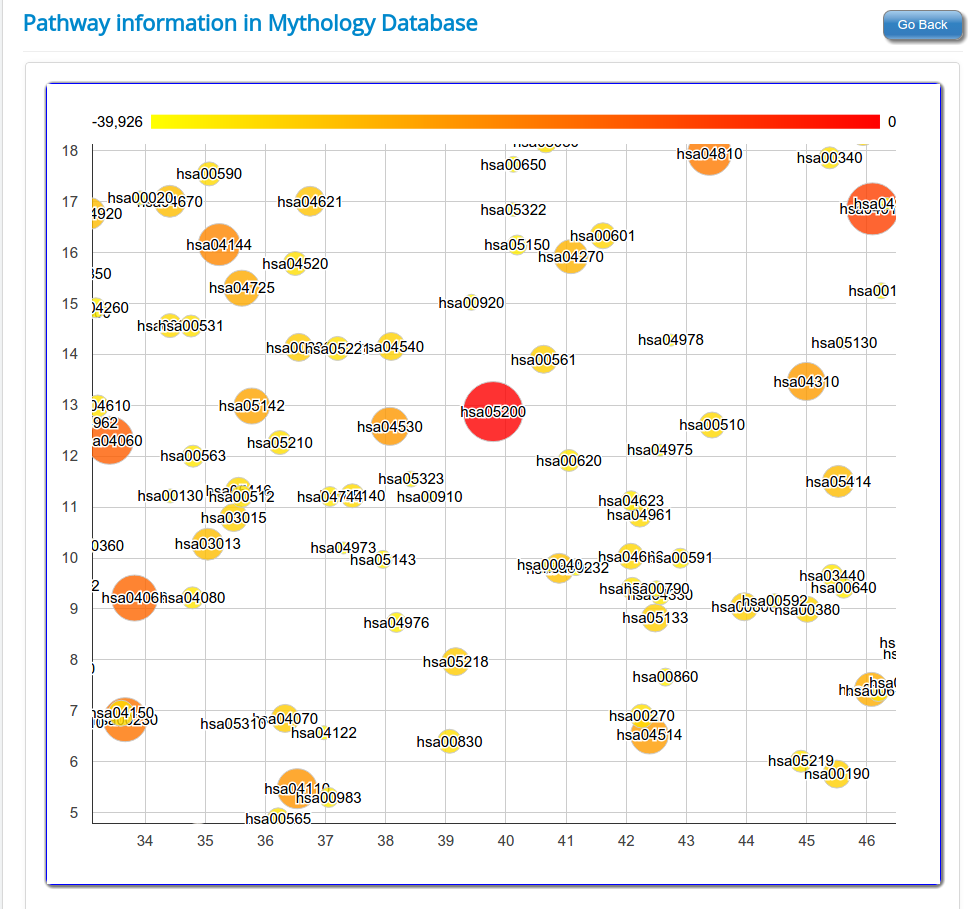
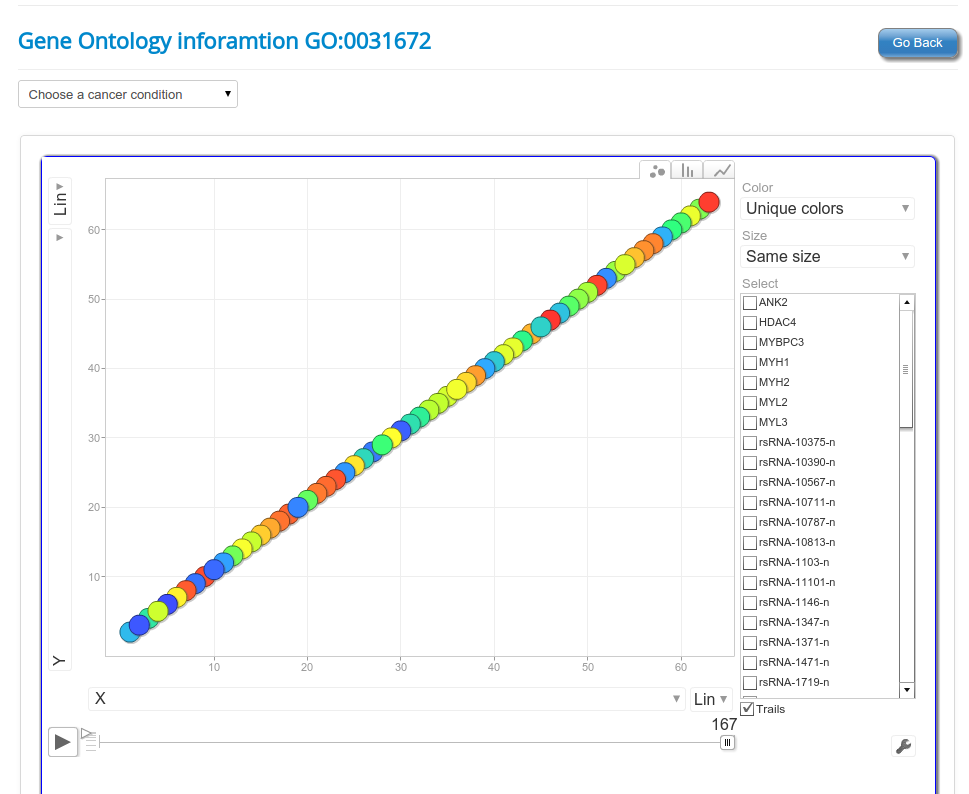
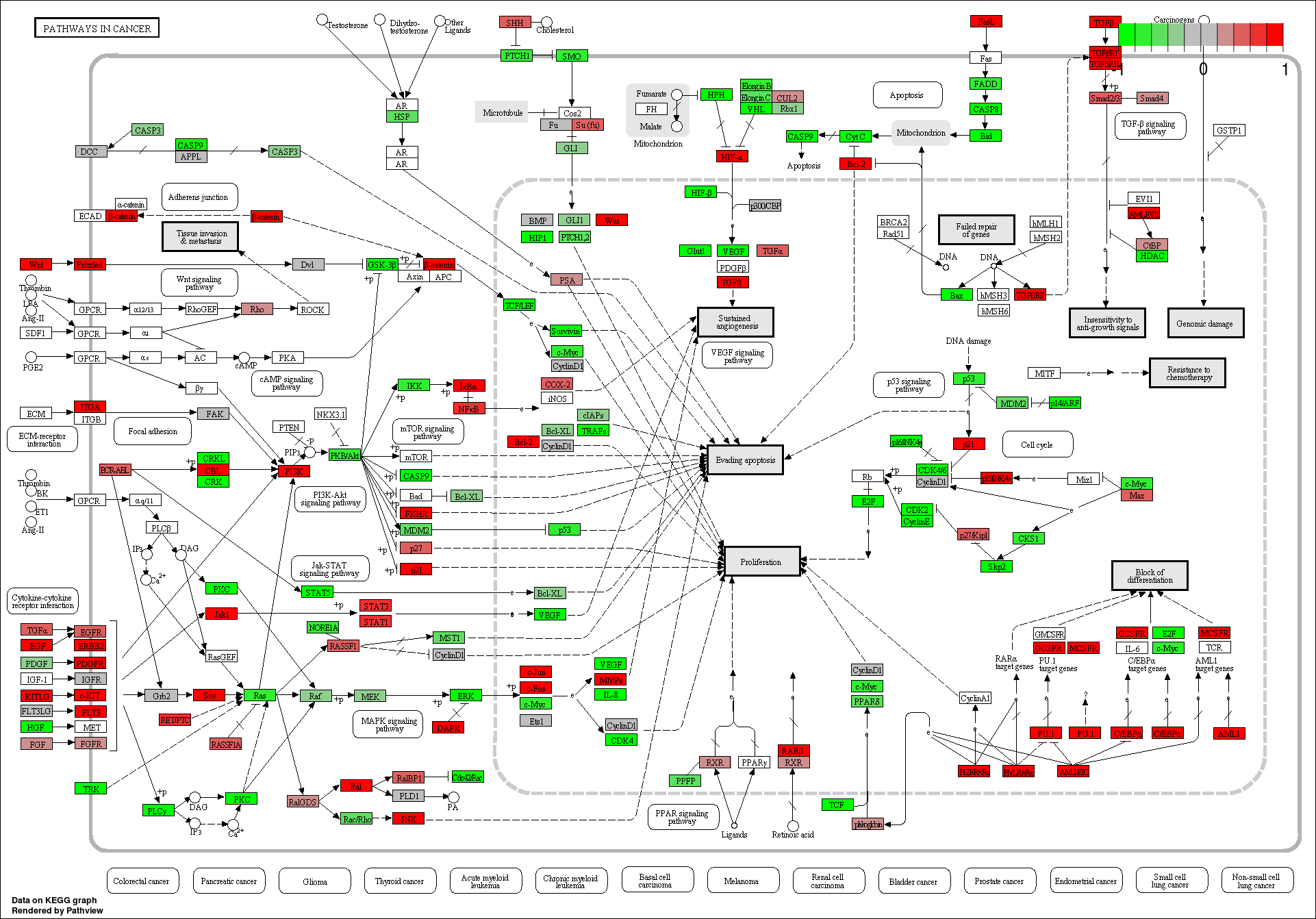
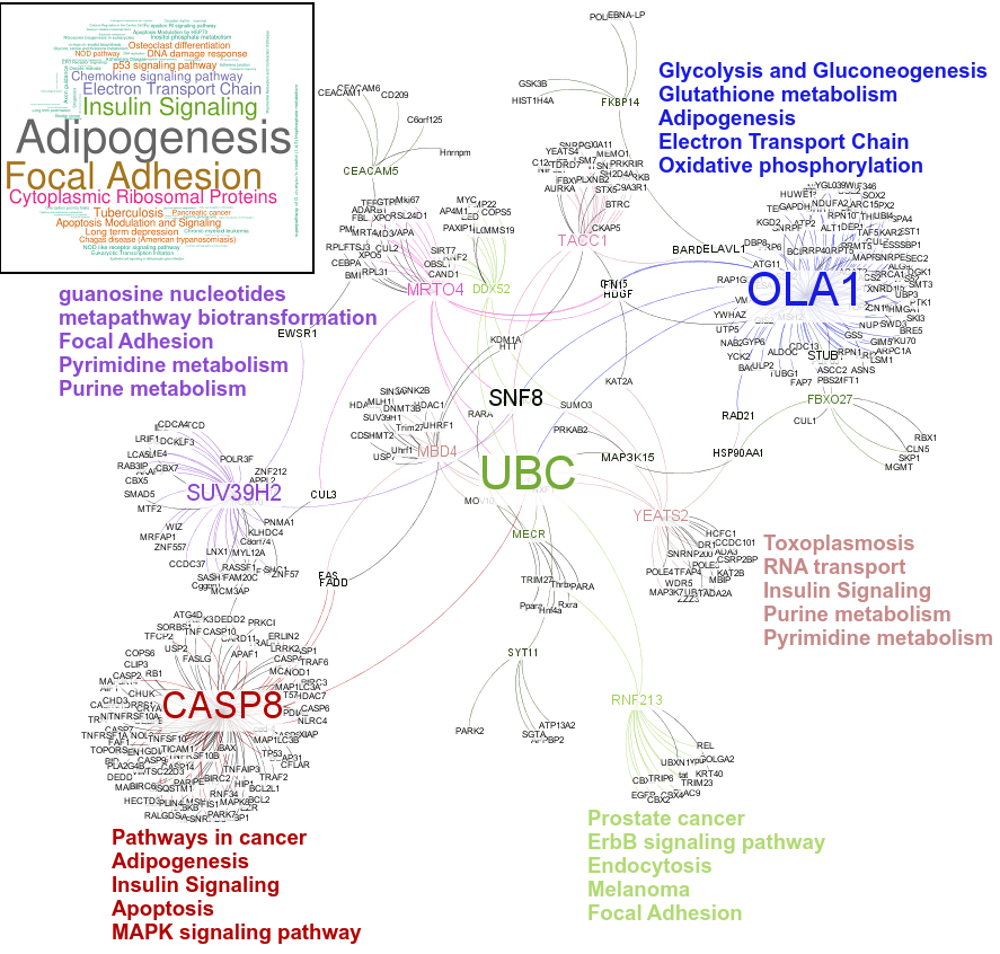
These novel rsRNAs (11,234 in this study) were found to regulate many important biological pathways. Related to cell growth and development and pathways which decide the fate of the cell converting from normal to tumor cell. Some of the pathways include pathways in cancer (hsa05200); Apoptosis (hsa04210) ; Cell cycle (hsa04110); p53 signaling pathway (hsa04115) and many more important pathways.
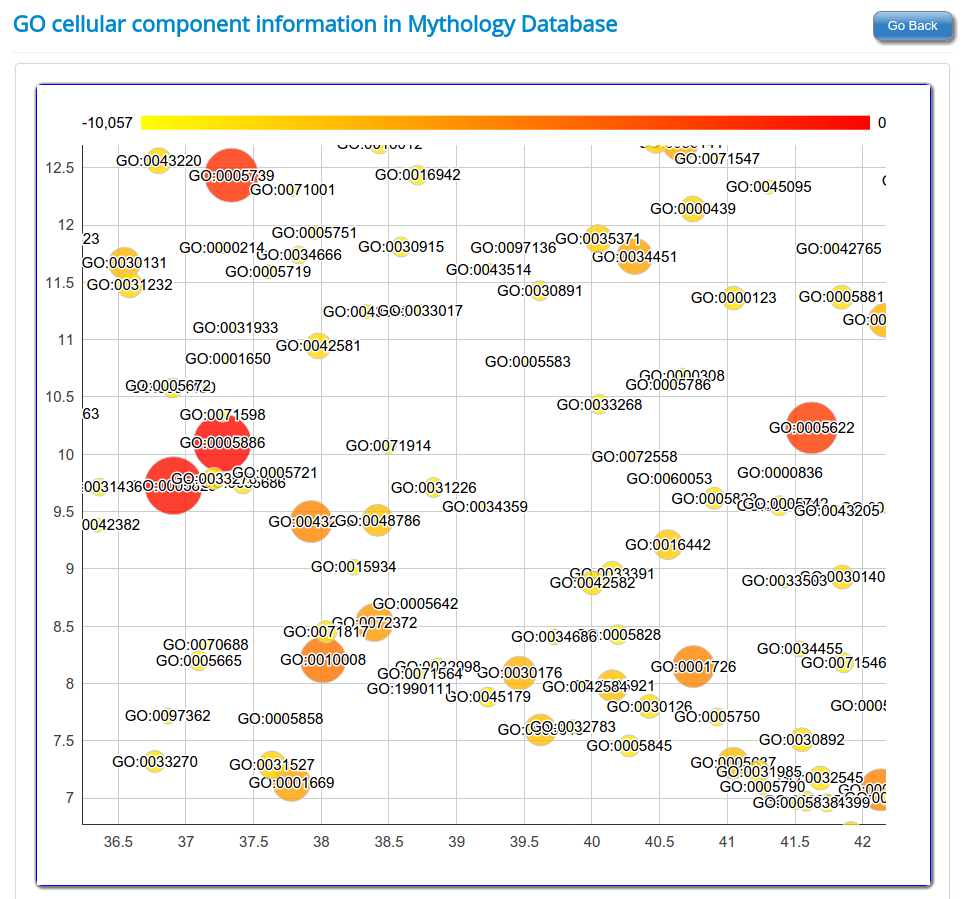
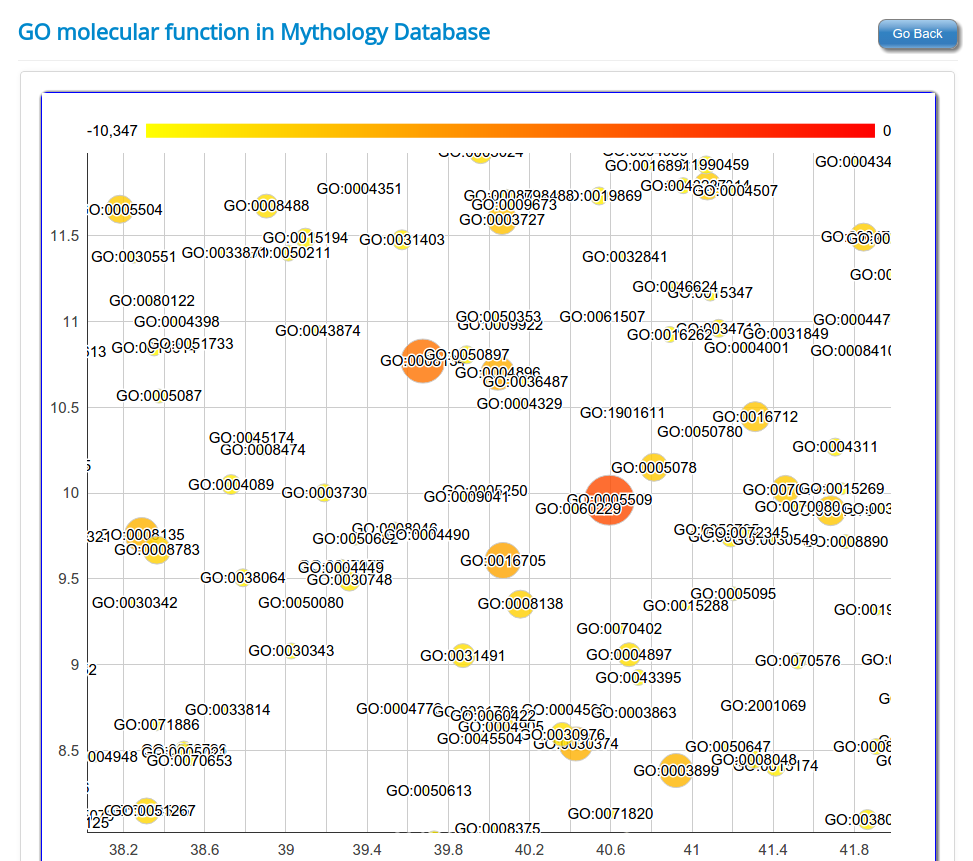
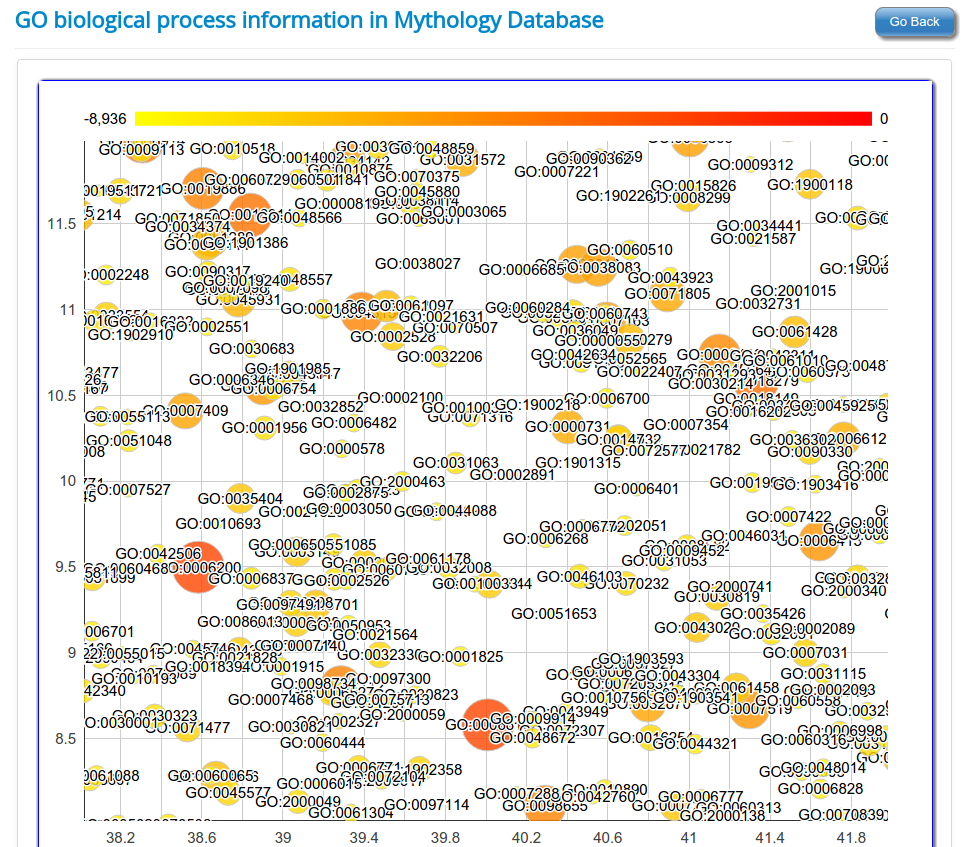
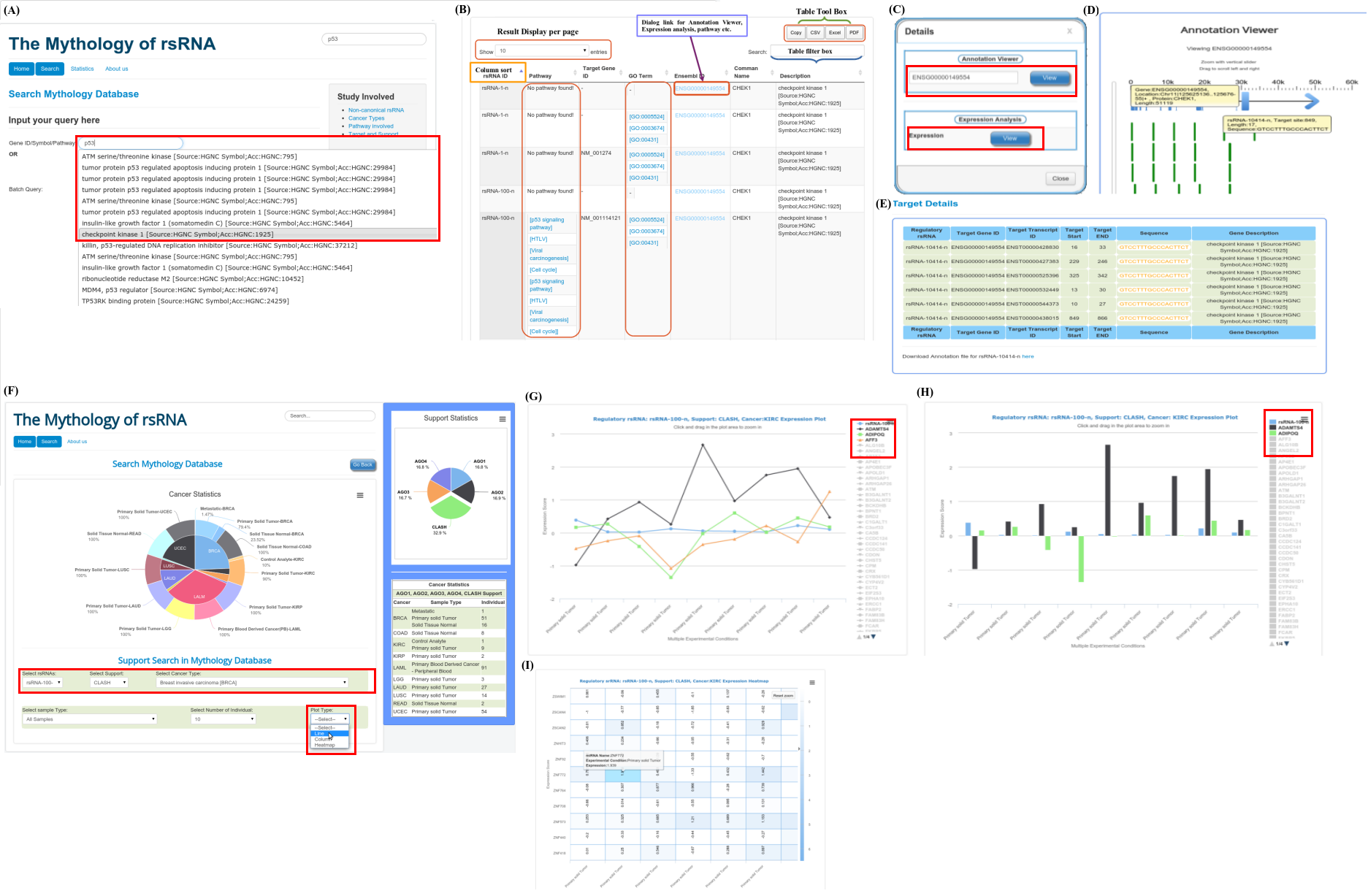
Gene Ontology based functional classification of rsRNAs
The identified rsRNAs were classified to regulate functions based on the Gene Ontology representation of respective target genes in each of the three GO category namely
Molecular function,
Biological process and,
Cellular component.
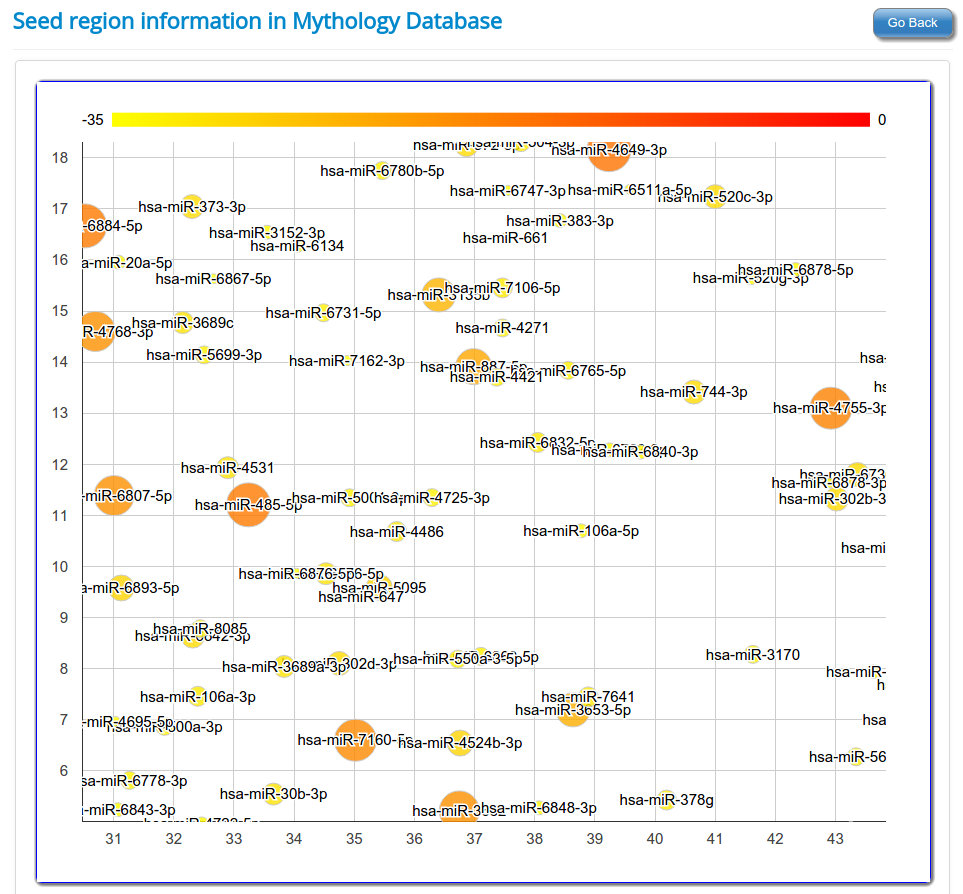
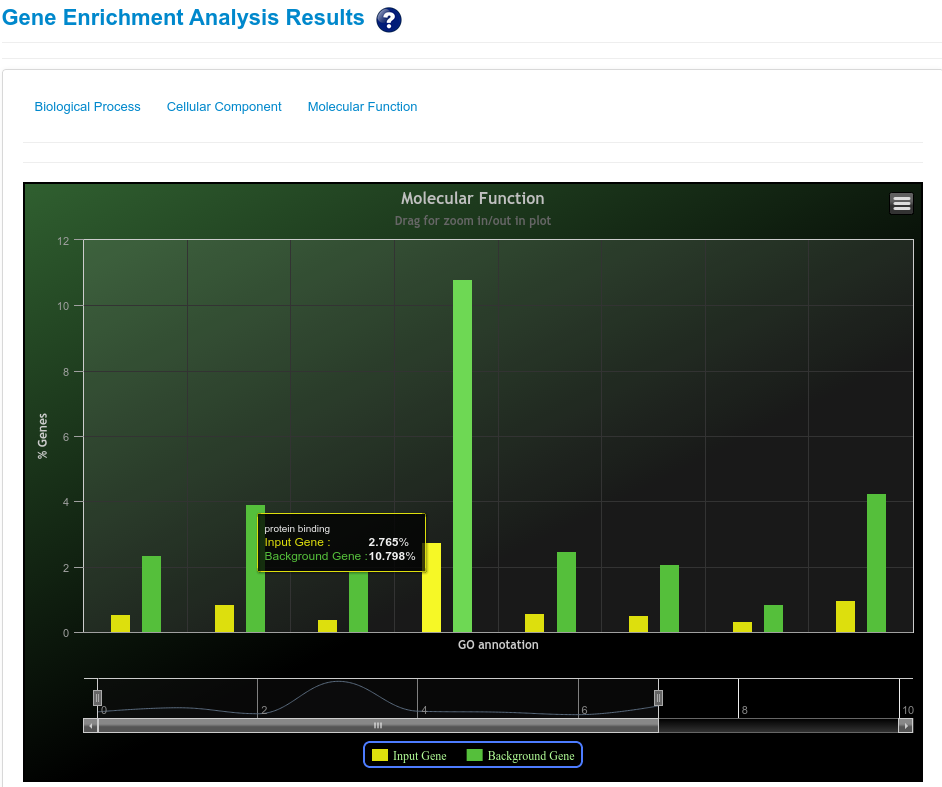
Seed region in mature miRNA play an important role in target gene identification and target binding. It was shown that seed region binds to the target site with either perfect complementarity or with near perfect complementarity. Mature miRNAs were classified in families according to conserved seed region and it was known that conserved seed region members target same or similar genes. Thus, to identify such similar sRNAs which have conserved seed region with already reported known mature miRNAs another clustering was performed for rsRNAs, based on seed region derived from known mature miRNA. A tree representation of rsRNAs shows how conserved is the seed region of rsRNAs with known mature miRNAs.
In humans and other animal species, the seed region of mature miRNAs starts from 2 nt till 8 nt. For this analysis, seed region of 11,234 novel rsRNAs and known mature miRNAs were extracted which corresponds to 6,371 unique seed sequences. Out of total 11,234 rsRNAs, most rsRNAs (411) share conserved seed region with “hsa-miR-7111-5p” with maximum of two mismatches. All the 11,234 rsRNAs shows conservation with known seed region of mature miRNAs at two mismatches where as 10,598 rsRNAs shows conservation with mature miRNAs at one mismatches. 2,353 rsRNAs align perfectly with seed region of mature miRNAs. The analysis shows that these rsRNAs have the ability to target the similar genes which were regulated by known mature miRNAs but were left undetected form the studies due to conventional practices of mature miRNA identification.
Using seed region analysis, form tree or seed region cluster, gene enrichment analysis can be performed for gene ontology categories namely Molecular function; Biological process and Cellular component. Gene enrichment analysis for rsRNAs targets helps to understand the preference for target genes by these common seed sharing rsRNAs.
Related Resources
Statistics
Browse rsRNAs
Visualization Tutorials
Contact us
Studio of Computational Biology & Bioinformatics,
Biotech Division,
CSIR-Institute of Himalayan Bioresource Technology,
Palampur 176061 (Himachal Pradesh), India
Email:ravish@ihbt.res.in
Website:http://scbb.ihbt.res.in/